Abstract
The deubiquitinase cylindromatosis (CYLD) functions as a tumor suppressor inhibiting cell proliferation in many cancer types including melanoma. Here we present evidence that a proportion of melanoma cells are nonetheless addicted to CYLD for survival. The expression levels of CYLD varied widely in melanoma cell lines and melanomas in vivo, with a subset of melanoma cell lines and melanomas displaying even higher levels of CYLD than melanocyte lines and nevi, respectively. Strikingly, although short hairpin RNA (shRNA) knockdown of CYLD promoted, as anticipated, cell proliferation in some melanoma cell lines, it reduced cell viability in a fraction of melanoma cell lines with relatively high levels of CYLD expression and did not impinge on survival and proliferation in a third type of melanoma cell lines. The decrease in cell viability caused by CYLD knockdown was due to induction of apoptosis, as it was associated with activation of the caspase cascade and was abolished by treatment with a general caspase inhibitor. Mechanistic investigations demonstrated that induction of apoptosis by CYLD knockdown was caused by upregulation of receptor-interacting protein kinase 1 (RIPK1) that was associated with elevated K63-linked polyubiquitination of the protein, indicating that CYLD is critical for controlling RIPK1 expression in these cells. Of note, microRNA (miR) profiling showed that miR-99b-3p that was predicted to target the 3′-untranslated region (3′-UTR) of the CYLD mRNA was reduced in melanoma cell lines with high levels of CYLD compared with melanocyte lines. Further functional studies confirmed that the reduction in miR-99b-3p expression was responsible for the increased expression of CYLD in a highly cell line-specific manner. Taken together, these results reveal an unexpected role of CYLD in promoting survival of a subset of melanoma cells and uncover the heterogeneity of CYLD expression and its biological significance in melanoma.
Key words: Cylindromatosis (CYLD), Melanoma, miR-99b-3p, Receptor-interacting protein kinase 1 (RIPK1)
INTRODUCTION
Treatment with small-molecule inhibitors against mutant BRAF and MEK, alone or in combination, has become the standard of care in patients with late-stage mutant BRAF melanomas1. Immunotherapeutic antibodies against the immune checkpoints T-lymphocyte-associated antigen 4 (CTLA-4) and programmed cell death protein 1 (PD-1) and the modified oncolytic herpes virus talimogene laharparepvec (T-VEC) have also been clinically available for treating the disease2,3. These approaches have improved outcomes of patients4. However, intrinsic and acquired resistance remains a major obstacle in the curative treatment of metastatic melanoma5,6. This is closely associated with genetic and epigenetic heterogeneity of melanoma cells7–11.
Although mutation of the gene encoding cylindromatosis (CYLD) was initially identified as a genetic predisposition for the development of the skin appendage tumors, cylindromas, and trichoepitheliomas12, it is now known that CYLD functions as a tumor suppressor in a variety of tissues, and its loss and mutation are associated with many types of cancer including melanoma13–15. As a deubiquitinase, CYLD removes K63-linked polyubiquitin chains from target proteins, leading to their degradation by the proteasomal system16. These include TNF receptor-associated factor 2 (TRAF2) and nuclear factor (NF)-κB essential modulator (NEMO) that are required for classic activation of NF-κB, and Bcl-3 that is necessary for noncanonical activation of NF-κB and has been shown to promote melanoma cell proliferation through upregulating cyclin D117,18. Moreover, CYLD has also been reported to promote melanoma tumorigenesis and metastasis through suppression of JNK/activating protein 1 (AP-1), leading to decreased expression of cyclin D1 and N-cadherin19.
CYLD can also negatively regulate the expression of receptor-interacting protein kinase 1 (RIPK1) that is emerging as an important determinant of cell fate in response to cellular stress through its deubiquitinase activity20. Past studies have shown that RIPK1 expression is upregulated and has an oncogenic role through promoting cell proliferation in melanoma21. However, overexpression of RIPK1 induces apoptosis in a proportion of melanoma cells and melanocytes21. Indeed, RIP1 overexpression triggers apoptosis in many other cell types through activating the caspase cascade22. It seems that the expression of RIP1 must be controlled tightly so that its cellular level remains constantly below a threshold to ensure avoidance of apoptosis21.
In this study, we have revisited the role of CYLD in melanoma cell survival and proliferation. We report here that CYLD expression levels are highly heterogenous among melanoma cell lines and melanomas in vivo and that its effect on the viability of melanoma cells also varied widely. We demonstrate that CYLD is necessary for survival of a subset of melanoma cells through controlling RIPK1 expression and that the reduction in microRNA-99b-3p (miR-99b-3p) expression is responsible for the increased CYLD expression in a cell line-specific manner.
MATERIALS AND METHODS
Cell Lines and Human Tissues
The human melanoma cell lines described previously were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% fetal calf serum (FCS)23. Human melanocytes (HEMn-MP) were purchased from Banksia Scientific and cultured as previously described24. Human fresh melanoma isolates were prepared from surgical specimens according to the published method25. All cell lines were verified to be free of mycoplasma contamination every 3 months and were authenticated using short tandem repeat (STR) profiling by the Australia Genome Research Facility (AGRF) in April 2018. Formalin-fixed paraffin-embedded (FFPE) melanocytic tumor tissues were retrieved from the Department of Tissue Pathology of Henan Provincial People’s Hospital. Studies using human tissues were approved by the Human Research Ethics Committee of Henan Provincial People’s Hospital, China.
Small Interference RNAs (siRNAs) and Plasmids
siRNAs were purchased from GenePharma (Shanghai, China) and transfected using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA). The siRNA sequences are as follows: CYLD siRNA1, 5′-GGGAAGUAUAGGACAGUAUTT-3′; CYLD siRNA2, 5′-GGUUCAUCCAGUCAUAAUATT-3′; has-miR-99b-3p mimic, 5′-CAAGCUCGUGUCUGUGGGUCCG-3′; anti-has-miR-99b-3p, 5′-CGGACCCACAGACACGAGCUUG-3′; RIPK1 siRNA1, 5′-CCUUCUGAGCAGCUUGAUUTT-3′; RIPK1 siRNA2, 5′-GCCAGCUGCUAAGUACCAATT-3′.
Flag-HA-CYLD (#22544), Flag-HA-GFP (#22612), and pRK5-HA-ubiquitin-K63 (#17606) were purchased from Addgene (Watertown, MA, USA). RIPK1 cDNA (pCMV-Myc-RIP1) and the control vector were purchased from Origene (Rockville, MD, USA)21. Reporter plasmids were constructed based on the psiCHECK-2 plasmid (Promega, San Luis Obispo, CA, USA). The primer sequences are as follows: RT-miR-99b-3p, 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCGGACCC-3′; miR-99b-3p-F, 5′-GTTCAAGCTCGTGTCT-3′; miR-99b-3p-R, 5′-CAGTGCGTGTCGTGGAGT-3′; CYLD 3′-UTR-HAF, 5′-AGTAATTCTAGGCGATCGAAGGTTTTATACTGCTAAGTGCTTGGTT-3′; CYLD 3′-UTR-HAR, 5′-TATTTTATTGCGGCCACACAGAGAAACAGTATGATAAACATCCA-3′; CYLD 3′-UTR 1659 MUT-F, 5′-AATGAAGAAATGGGTGAATGTCGTTGTCAATGT-3′; CYLD 3′-UTR 1659 MUT-R, 5′-TTTTAAAATCACATTGACAAGCTCATTCACCCA-3′; CYLD 3′-UTR 2549 MUT-F, 5′-GCTGGGCTGCATGGCACAGGTCGTTATGTGCCT-3′; CYLD 3′-UTR 2549 MUT-R, 5′-AATAACCAGCAGGCACATAAGCTCCTGTGCCAT-3′; CYLD 3′-UTR 3602 MUT-F, 5′-GCTCTGTACACTTAGACAACTCGTTGACCTCTT-3′; CYLD 3′-UTR 3602 MUT-R, 5′-ACTAAAGCTCAAGAGGTCAAGCTGTTGTCTAAG-3′; CYLD-qPCR-F, 5′-TGGGATGGAAGATTTGATGGAG-3′; CYLD-qPCR-R, 5′-CATAAAGGCAAGTTTGGGAGG-3′.
RIPK1 Ser320Asp and Ser25Asp mutation plasmids were constructed based on the pCMV-Myc-RIP1 plasmid (Origene). The primer sequences are as follows: RIPK1 Ser25Asp-F, 5′-AGAACTGGACGATGGAGGCTTTGGGAAGGTGTC-3′; RIPK1 Ser25Asp-R, 5′-CAAAGCCTCCATCGTCCAGTTCTGCACTCTCCA-3′; RIPK1 Ser320Asp-F, 5′-GAGAATGCAGGATCTTCAACTTGATTGTGTGGC-3′; RIPK1 Ser320Asp-R, 5′-CAAGTTGAAGATCCTGCATTCTCTTCACAACTG-3′.
Lentiviral Gene Transduction
MISSION human short hairpin RNA (shRNA) lentiviral transduction particles targeting CYLD (TRCN0000218454) and control (SHC002V) particles were purchased from Sigma-Aldrich (St. Louis, MO, USA). CYLD cDNA lentiviral particles were transducted as described previously23. In brief, melanocyte cells were seeded at approximately 80% confluency 1 day ahead. Melanocytes were transduced and incubated with lentiviral particles for 48 h.
Immunohistochemistry (IHC)
IHC staining and quantitation were performed as described previously24. Briefly, antigen retrieval was performed in a pressure cooker for 30 s at 125°C. Antibody detection was performed using the Dako Envision HRP Detection system/DAB as per the manufacturer’s instructions. Slides were counterstained with Azure B and hematoxylin24. The antibody against CYLD (PA5-34630) was purchased from Thermo Fisher Scientific (Scoresby, VIC, Australia).
Western Blotting
Western blotting was carried out and protein bands were quantitated by ImageJ as described previously26. The antibody against CYLD (ab137524) was purchased from Abcam (Cambridge, UK). The antibody against HA-tag (#2367) was purchased from Cell Signaling Technology (Beverly, MA, USA). The antibody against caspase 3 (AAP-113) and caspase 8 (AAM-118) were from Enzo Life Sciences (Dural, NSW, Australia). The antibodies against PARP (556494), c-IAP-1 (556533), c-IAP-2 (552782), and RIPK1 (51-6559GR) were from BD Biosciences (North Ryde, NSW, Australia). The antibodies against cyclin D1 (sc-753) and FADD (sc-2717488) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Cell Viability
Cell viability was quantitated using the CellTiter-Glo Luminescent Cell Viability Assay Kit (G7570; Promega) as described previously27. Luminescence was recorded by a Synergy 2 multidetection microplate reader (BioTek, Winooski, VT, USA).
Clonogenic Assays
Cells were seeded at 2 × 105 per well onto six-well culture plates and allowed to grow for 16–24 h followed by transfection. Cells were then reseeded at 1,000 per well onto six-well culture plates and allowed to grow for a further 14 days before fixation with methanol and staining with crystal violet (0.5%). Colony area was quantitated using ImageJ plugin.
Cell Cycle Analysis
Cells were seeded at 4 × 104 per well onto 24-well plates and allowed to grow for 16–24 h followed by transfection. Cells were fixed and followed by propidium iodide staining as described previously28.
Apoptosis
Apoptotic cells were detected as described previously23.
Cell Proliferation Assays
5-Bromo-2′-deoxyuridine (BrdU) cell proliferation assays were carried out using an assay kit as per the manufacturer’s instructions (#6813; Cell Signaling Technology). Briefly, cells were seeded at 8 × 103 cells/well in 96-well plates overnight before treatment as described previously21. BrdU (10 mmol/L) was added and cells were incubated for 4 h before BrdU assays were carried out. Absorbance was read at 450 nm using a Synergy 2 multidetection microplate reader.
Immunoprecipitation
Immunoprecipitation was carried out as described previously24. In brief, extracts were mixed and precipitated with antibody (RIPK1; 51-6559) and protein A/G Agarose beads by incubation at 4°C. The bound proteins were removed by boiling in sodium dodecyl sulfate (SDS) buffer and resolved in SDS-polyacrylamide gel electrophoresis (PAGE) gels for immunoblotting analysis.
Luciferase Reporter Assay
Reporter activities were measured as per the manufacturer’s protocol (Dual-Glo® Luciferase Assay System; Promega). Briefly, psiCHECK-2 vector-based reporter plasmids were cotransfected with miRNA mimics or anti-miRNAs into cells. Firefly and Renilla luciferase activities were recorded using a microplate reader (BioTek), respectively.
Statistical Analysis
Statistical analysis was carried out using GraphPad Prism 8. Statistical significance was analyzed by Student’s t-test expressed as a p value. A value of p < 0.05 was considered as statistically significant.
RESULTS
Heterogeneous Expression of CYLD in Human Melanoma Cells
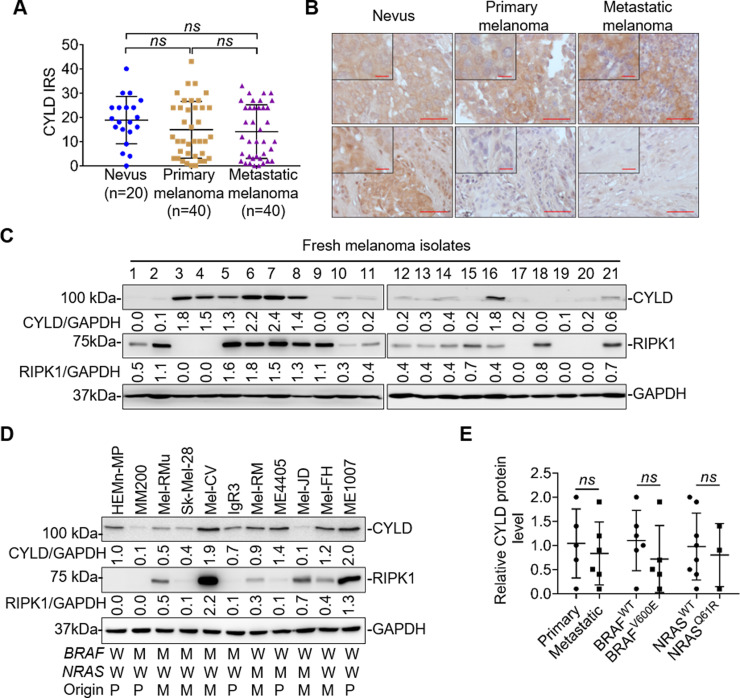
We examined the expression of CYLD in relation to melanoma development and progression using IHC in 100 FFPE melanocytic tumors. The results showed that although there was tendency for melanomas to express lower levels of CYLD than nevi, the difference was not statistically significant (Fig. 1A). Similarly, there was no significant difference in CYLD expression levels between primary and metastatic melanomas. Of note, the levels of CYLD in melanomas and nevi varied considerably (Fig. 1A and B). In particular, a proportion of melanomas expressed even higher levels of CYLD compared with the average level in nevi (Fig. 1A and B). In accordance, examination of 21 fresh metastatic melanoma isolates by Western blotting showed that there were wide variations in CYLD expression levels (Fig. 1C).
Figure 1.
Cylindromatosis (CYLD) is heterogeneously expressed in human melanocytic cells. (A) Quantification of CYLD expression in melanocytic tumors. Data shown are mean immunoreactive score (IRS) ± standard error of the mean (SEM). ns, p > 0.05, Kruskal–Wallis test. (B) Representative microphotographs of immunohistochemistry (IHC) staining of CYLD on melanocytic tissue sections. Scale bar: 100 μm. (C, D) Whole-cell lysates from melanocytes and melanoma cells were subjected to Western blotting. Data shown are representative of three individual experiments. W, wild-type; M, BRAFV600E or NRASQ61R mutation; P, primary melanoma; M, metastatic melanoma. (E) The relative abundance of CYLD was compared between primary (n = 5) and metastatic (n = 6), BRAFWT (n = 6) and BRAFV600E (n = 5), and NRASWT (n = 8) and NRASQ61R (n = 3) melanoma cell lines. Data were adapted from (D). ns, p > 0.05, Student’s t-test.
We also tested CYLD expression in a panel of 10 melanoma cell lines, which were generated from melanomas of different stages and had varying status of the most common mutations in BRAF (BRAFV600E) and NRAS (NRASQ61R) (Fig. 1D). All the melanoma cell lines carried wild-type CYLD gene as tested using exon sequencing. CYLD was similarly expressed at various levels in melanoma cell lines that were not associated with their origins and genetic backgrounds (Fig. 1E). While 6 of the 10 melanoma cell lines displayed lower levels of CYLD compared with the melanocyte line HEMn-MP, the other 4 melanoma lines (Mel-CV, ME4405, Mel-FH, and ME1007) exhibited higher or comparable levels of CYLD relative to HEMn-MP. Collectively, these results revealed that the expression levels of CYLD are highly heterogenous in melanoma cells29.
CYLD Differentially Regulates Melanoma Cell Survival and Proliferation
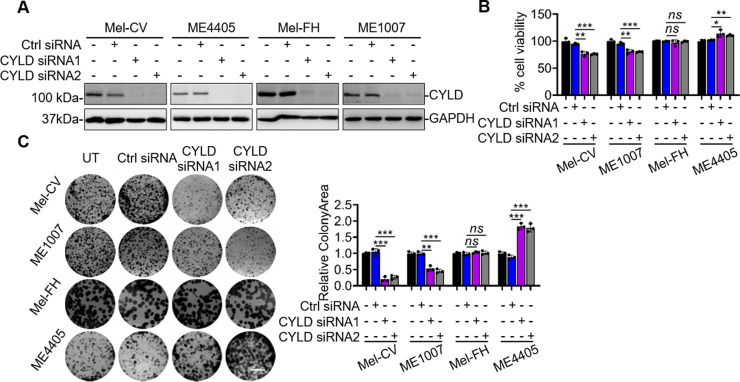
We investigated the functional significance of the relatively high expression CYLD in Mel-CV, Mel-FH, ME4405, and ME1007 cells by siRNA silencing of CYLD (Fig. 2A). Strikingly, while CYLD silencing decreased cell viability as measured using CellTiter-Glo assays in Mel-CV and ME1007 cells, it resulted in a moderate yet statistically significant increase in cell viability in ME4405 cells and had no effect on the viability of Mel-FH cells (Fig. 2B). These varying effects of CYLD on cell viability in different melanoma cell lines were more prominently reflected in clonogenic assays, where CYLD silencing reduced the clonogenic potential in Mel-CV and ME1007 cells but promoted the clonogenicity in ME4405 cells. On the other hand, it did not impinge on colony formation in Mel-FH cells (Fig. 2C).
Figure 2.
CYLD differentially regulates melanoma cell survival and proliferation. (A, B) Mel-CV, ME4405, Mel-FH, and ME1007 cells transfected with indicated CYLD short hairpin RNAs (siRNAs) were subjected to Western blotting (A) and CellTiter-Glo assays (B). Data shown are representative of three individual experiments (A) or means ± SEM (B). *p < 0.05; **p < 0.01; ***p < 0.001, Student’s t-test. (C) Mel-CV, ME1007, Mel-FH, and ME4405 cells transfected with indicated CYLD siRNAs were subject to clonogenic assays (left). Scale bar: 1 cm. Relative colony area were measured by ImageJ (right). Colony area in control cells was arbitrarily designated as 1. Data shown are representative of three individual experiments (left) or means ± SEM (right). n = 3. **p < 0.01; ***p < 0.001, Student’s t-test.
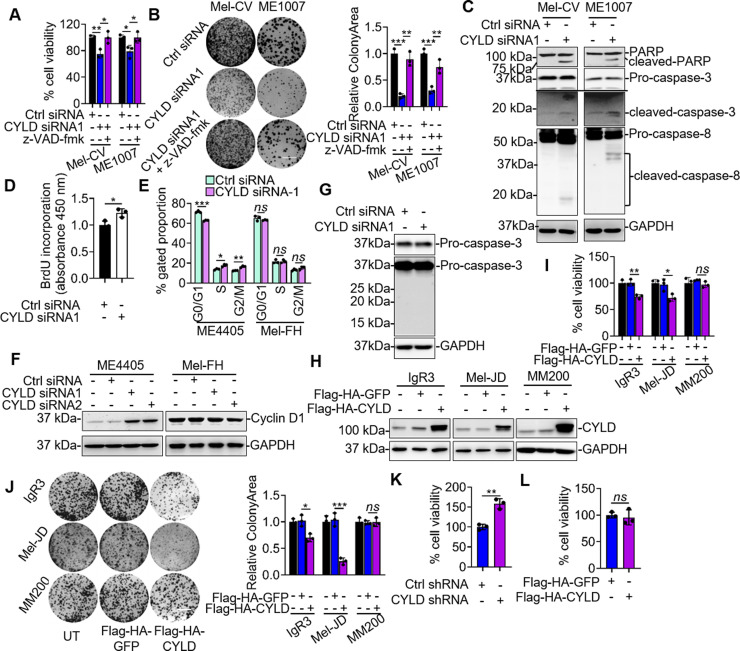
We focused on investigation of the mechanism by which CYLD promotes cell survival in Mel-CV and ME1007 cells. The inhibitory effect of CYLD silencing on the viability of Mel-CV and ME1007 cells was due to induction of apoptosis, as the addition of the general caspase inhibitor z-VAD-fmk efficiently rescued the cells (Fig. 3A and B). Consistently, silencing of CYLD caused activation of caspase 3 and cleavage of poly(ADP-ribose) polymerase (PARP) (Fig. 3C). Moreover, it triggered activation of caspase 8 (Fig. 3C), suggesting that caspase 8 is involved in apoptosis when CYLD is silenced in these cells30. The promoting effect of CYLD silencing on ME4405 cell viability was due to increased cell proliferation as shown by increased incorporation of BrdU (Fig. 3D). This was associated with a decrease in the fraction of cells in the G1/G0 phase, an increase in the proportion of cells in the S phase and G2/M phase, and upregulation of cyclin D1 expression (Fig. 3E and F)14. In contrast, silencing of CYLD did not have any effect on caspase activation, cell cycle progression, and cyclin D1 expression in Mel-FH cells (Fig. 3E–G). Collectively, these results suggest that CYLD differentially regulates melanoma cell survival and proliferation in a cell line-dependent manner. In support, overexpression of CYLD in IgR3 and Mel-JD cells that expressed relatively low levels of endogenous CYLD resulted in inhibition of cell proliferation (Fig. 3H–J). However, overexpression of CYLD in MM200 cells that similarly displayed low endogenous levels of CYLD did not significantly impinge on cell viability (Fig. 3H–J). Of note, while silencing of CYLD promoted proliferation of melanocytes (Fig. 3K), overexpression of CYLD did not have any significant effect on melanocyte growth (Fig. 3L).
Figure 3.
CYLD differentially regulates melanoma cell survival and proliferation in a cell line-dependent manner. (A, B) Mel-CV and ME1007 cell lines transfected with indicated siRNAs and with or without z-VAD-fmk were subject to CellTiter-Glo assays (A) and clonogenic assay (B). Data shown are representative of three individual experiments or means ± SEM. n = 3. *p < 0.05; **p < 0.01; ***p < 0.001, Student’s t-test. (C) Whole-cell lysates from Mel-CV and ME1007 transfected with indicated siRNAs were subjected to Western blotting. Data shown are representative of three individual experiments. (D) ME4405 cells transfected with indicated siRNA were subjected to 5-bromo-2′-deoxyuridine (BrdU) incorporation assays. Data shown are representative of three individual experiments or means ± SEM. n = 3. *p < 0.05. (E, F) ME4405 and Mel-FH cells transfected with indicated siRNAs were subjected to cell cycle distribution (E) and Western blotting (F). Data shown are representative of three individual experiments or means ± SEM. n = 3. *p < 0.05; **p < 0.01; ***p < 0.001; ns, p > 0.05, Student’s t-test. (G) Mel-FH cells were transfected with indicated siRNA and subjected to Western blotting. Data shown are representative of three individual experiments. (H–J) IgR3, Mel-JD, and MM200 cell lines transfected with indicated plasmids were subjected to Western blotting (H), CellTiter-Glo assays (I), and clonogenic assay (J). Scale bar: 1 cm. Data shown are representative of three individual experiments or means ± SEM. n = 3. *p < 0.05; **p < 0.01; ***p < 0.001; ns, p > 0.05, Student’s t-test. (K, L) HEMn-MP melanocytes transduced with indicated shRNAs (K) or cDNA (L) were subjected to CellTiter-Glo assays. Data shown are means ± SEM. n = 3. **p < 0.01; ns, p > 0.05, Student’s t-test.
CYLD Is Critical for Protection Against the Apoptosis-Inducing Potential of RIPK1 in a Subset of Melanoma Cells
We next examined the mechanism responsible for induction of apoptosis in Mel-CV and ME1007 cells by CYLD silencing. As a K63-specific deubiquitinase, CYLD removes K63-linked polyubiquitin chains from RIPK1, thus promoting its degradation20,31. Since overexpression of RIPK1 in melanoma cells triggers apoptosis21, we examined the potential involvement of RIPK1 in induction of apoptosis upon CYLD knockdown in Mel-CV and ME1007 cells. Silencing of CYLD caused upregulation of RIPK1 in Mel-CV, ME1007, Mel-FH, and ME4405 cells regardless of their differential apoptotic responses (Fig. 4A). This was associated with an increase in K63-linked polyubiquitination of RIPK1 (Fig. 4B). In contrast, overexpression of CYLD caused downregulation of RIPK1 in Mel-CV and ME1007 cells, which was nevertheless reversed by the treatment with the proteasome inhibitor MG132 (Fig. 4C), substantiating that downregulation of RIPK1 expression by CYLD is mediated by the proteasomal degradation20,30,32,33. Cosilencing of RIPK1 diminished apoptosis induced by CYLD silencing in Mel-CV and ME1007 cells, substantiating the role of RIPK1 in induction of apoptosis by silencing of CYLD. However, cosilencing of RIPK1 had no significant effect on CYLD silencing-triggered increase in proliferation of ME4405 cells and did not impinge on proliferation of Mel-FH cells with CYLD silenced (Fig. 4D and E). Together, these results indicate that the increased expression of RIPK1 is responsible for induction of apoptosis in Mel-CV and ME1007 cells with CYLD silenced and that it does not play a role in the promotion of proliferation by CYLD silencing in ME4405 cells. Of note, the constitutive levels of RIPK1 expression were markedly higher in Mel-CV and ME1007 cells than in ME4405 and Mel-FH cells (Fig. 1D). Moreover, the overall levels of RIPK1 after CYLD silencing were higher in Mel-CV and ME1007 cells than in ME4405 and Mel-FH cells (Fig. 4A). Similar to CYLD expression levels, RIPK1 expression levels varied widely in fresh melanoma isolates (Fig. 1C). However, there is no significant relationship between CYLD and RIPK1 expression levels, suggesting that other mechanisms besides the expression of CYLD also participate in regulation of RIPK1 expression34,35.
Figure 4.
CYLD is critical for protection against the receptor-interacting protein kinase 1 (RIPK1)-induced apoptosis in a subset of melanoma cells. (A) Mel-CV, ME1007, Mel-FH, and ME4405 cell lines transfected with indicated siRNAs were subjected to Western blotting. Data shown are representative of three individual experiments. (B) Immunoprecipitates with RIPK1 antibody from Mel-CV and ME1007 cells transfected with indicated siRNAs and plasmids were subjected to Western blotting. Data shown are representative of three individual experiments. (C) Whole-cell lysates from Mel-CV and ME1007 cell lines transfected with indicated plasmids with or without treatment with MG132 were subjected to Western blotting. Data shown are representative of three individual experiments. (D) Mel-CV and ME1007 cells transfected with indicated siRNAs were subjected to propidium iodide (PI)/annexin V staining assays. Data shown are representative of three individual experiments (left) or means ± SEM (right). n = 3. ***p < 0.001, Student’s t-test. (E) ME4405 and Mel-FH cells transfected with indicated siRNAs were subjected to CellTiter-Glo assays. Data shown are means ± SEM. *p < 0.05; **p < 0.01; ns, p > 0.05, Student’s t-test. (F) Whole-cell lysates from Mel-CV and Mel-FH cells were subjected to Western blotting. Data shown are representative of three individual experiments. (G) Mel-CV and ME1007 cells with CYLD knocked down by siRNA were transfected with the indicated plasmids were subjected to PI/annexin V staining assays (top) and Western blotting (bottom). Data shown are means ±SEM (top) or representative of three individual experiments (bottom). n = 3. **p < 0.01; ***p < 0.001, Student’s t-test.
As Fas-associated protein with death domain (FADD), cellular inhibitor of apoptosis 1 (cIAP1), and cIAP2 are all known to regulate RIPK1-mediated apoptosis20,36, we compared the expression levels of FADD, cIAP1, and cIAP2 between Mel-FH and Mel-CV cells that displayed different sensitivity to apoptosis induced by silencing of CYLD. The results showed that there were no noticeable differences in FADD, cIAP1, and cIAP2 levels between these two cell lines (Fig. 4F), implicating that FADD, cIAP1, and cIAP2 do not play a major role in differentially regulating apoptosis in these cells when CYLD is silenced.
Since the proapoptotic function of RIPK1 can be prevented by phosphorylation of Ser320 or Ser2537,38, we introduced the RIPK1 mutant with Ser320 or Ser25 constitutively phosphorylated into Mel-CV and ME1007 cells to test whether it impinges on apoptosis induced by silencing of CYLD. The results showed that the expression of either of the mutants attenuated apoptosis in Mel-CV and ME1007 cells when CYLD was silenced (Fig. 4A and G), further consolidating the role of RIPK1 in apoptosis induced by silencing of CYLD in melanoma cells.
Cell Line-Dependent Regulation of CYLD by miR-99b-3p
Although CYLD expression at the protein level varied considerably in melanoma cell lines (Fig. 1C and D)29, its mRNA expression levels were generally lower in melanoma cell lines than the HEMn-MP melanocyte line (Fig. 5A). In particular, Mel-CV, ME4405, and Mel-FH that expressed relatively high levels of the CYLD protein exhibited even markedly lower levels of CYLD mRNA than HEMn-MP melanocytes (Fig. 5A). These results indicate that posttranscriptional upregulation is responsible for the increase in CYLD protein levels in these melanoma cell lines39,40. Noticeably, the turnover rates of the CYLD protein remained similar in Mel-CV, ME4405, Mel-FH, and HEMn-MP cells (Fig. 5B), suggesting that a translational increase is the main cause of CYLD upregulation in these melanoma cell lines.
Figure 5.
Cell line-dependent regulation of CYLD by microRNA-99b-3p (miR-99b-3p). (A) Total RNAs from melanocytes and melanoma cells were subjected to quantitative polymerase chain reaction (qPCR) analysis. Data shown are means ± SEM. (B) Mel-CV, ME4405, Mel-FH, and HEMn-MP cells were treated with or without cycloheximide (CHX; 5 μg/ml) for indicated periods. Whole-cell lysates were subjected to Western blotting (left). Quantification of CYLD relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is also shown (right). Mean ± SEM, n = 3, Student’s t-test. (C) Top 10 miRNAs that expressed the lowest level in melanoma cell lines Mel-CV and ME1007 in comparison with melanocytes. (D) Total RNAs from melanocytes and melanoma cells were subjected to qPCR analysis. Data shown are means ± SEM. (E) A schematic illustration of base pairing between miR-99b-3p and the 3′-untranslated region (3′-UTR) of CYLD. (F) A schematic illustration of psiCHECK2-based luciferase reporter constructs. (G) Luciferase reporter activity measured in Mel-CV and ME1007 cells after cotransfection with indicated reporter constructs and miRNA mimics. Data shown are means ± SEM, n = 3. *p < 0.05; ***p < 0.001, Student’s t-test. (H) Luciferase reporter activity measured in Mel-CV and ME1007 cells after cotransfection with indicated reporter constructs and anti-miRNA oligonucleotides. Data shown are means ± SEM. n = 3. **p < 0.01; ***p < 0.001, Student’s t-test. (I) Luciferase reporter activity measured in ME4405 and Mel-FH cells transfected with the indicated reporter constructs with or without cotransfection of miR-99b-3p mimics or anti-miR-99b-3p oligonucleotides. Data shown are means ± SEM. n = 3. p > 0.05, Student’s t-test. (J, K) Whole-cell lysates from Mel-CV and ME1007 cells transfected with indicated miRNA mimics (J) or anti-miRNA oligonucleotides (K) were subjected to qPCR analysis (top) and Western blotting (bottom). Data shown are representative of means ± SEM or three individual experiments. **p < 0.01; ***p < 0.001; ****p < 0.00001, Student’s t-test. (L) Whole-cell lysates from ME4405 and Mel-FH cells transfected with indicated miRNA mimics (top) or anti-miRNA oligonucleotides (bottom) were subjected to Western Blotting. Data shown are representative of three individual experiments.
We examined whether miRNAs that commonly target transcripts to block their translation and are often deregulated in cancer cells are involved in upregulation of CYLD in the subset of melanoma cell lines by comparing miRNA expression profiles between Mel-CV and ME1007 melanoma cell lines and the HEMn-MP melanocyte line (Fig. 5C)41. Among miRNAs that were differentially expressed, a decrease in miR-99b-3p in Mel-CV and ME1007 cells was the most pronounced (Fig. 5C). Subsequent quantitative polymerase chain reaction (qPCR) analysis showed that miR-99b-3p was commonly reduced in melanoma cell lines compared with the melanocyte line as shown in qPCR analysis (Fig. 5D). Interestingly, the 3′-untranslated region (3′-UTR) of the CYLD mRNA contained three regions largely complementary with the “seed” region of miR-99b-3p, one region matching seven bases (nucleotides 1,674–1,680) and another two regions matching six and five bases, respectively (nucleotides 3,602–3,623 and 2,549–2,572) (Fig. 5E).
To test whether miR-99b-3p targets the CYLD mRNA in melanoma cells, we introduced luciferase reporter plasmids of the 3′-UTR of CYLD into Mel-CV, ME1007, ME4405, and Mel-FH cells (Fig. 5F). The report activity was markedly suppressed by the presence of the 3′-UTR of CYLD in Mel-CV and ME1007, which was however reversed partially when the segment encompassing nucleotides 1,675–1,677, 2,567–2,569, and 3,618–3,620 of the 3′-UTR was mutated (Fig. 5G), suggesting that the 3′-UTR of CYLD was inhibited by endogenous miR-99b-3p. In accordance, cointroduction of anti-miR-99b-3p into Mel-CV and ME1007 cells increased (Fig. 5H), whereas the addition of miR-99b-3p mimics further reduced the reporter activity (Fig. 5G). Therefore, miR-99b-3p targets the 3′-UTR of the CYLD mRNA in Mel-CV and ME1007 cells. In contrast, there was no significant difference in the report activity in the presence or absence of the wild-type or mutated 3′-UTR of CYLD in ME4405 and Mel-FH cells (Fig. 5I). Moreover, cointroduction of anti-miR-99b-3p or miR-99b-3p mimics did not cause any change in the reporter activity, indicating that miR-99b-3p does not impinge on the 3′-UTR of the CYLD mRNA in these cells. Consistent with the results of luciferase assays, introduction of miR-99b-3p mimics downregulated, whereas introduction of anti-miR-99b-3p upregulated endogenous CYLD protein levels in Mel-CV and ME1007 cells (Fig. 5J and K). However, introduction of neither miR-99b-3p mimics nor anti-miR-99b-3p alters the levels in ME4405 and Mel-FH cells (Fig. 5L). Taken together, these results indicate that miR-99b-3p selectively targets the 3′-UTR of CYLD mRNA in melanoma cells in a cell line-dependent manner.
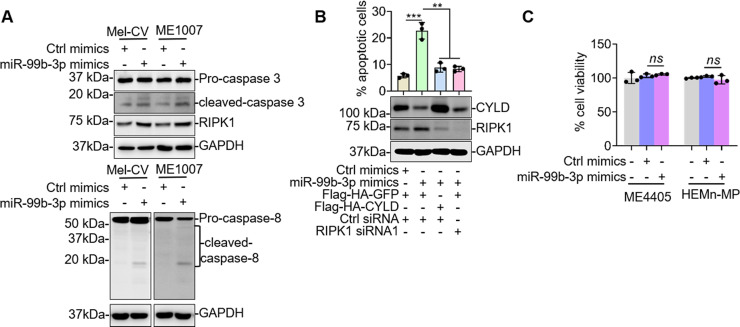
We also examined the functional significance of miR-99b-3p-mediated regulation of CYLD expression in Mel-CV and ME1007 cells. Introduction of miR-99b-3p mimics resulted in apoptosis that was associated with activation of caspase-3 and upregulation of RIPK1 (Fig. 6A), recapitulating the effect of CYLD knockdown (Fig. 3C). Furthermore, introduction of miR-99b-3p mimics caused activation of caspase 8, consistent with the notion that apoptosis induced by high expression of RIPK1 is mediated by caspase 830. Cointroduction of CYLD or RIPK1 siRNA abolished apoptosis induced by introduction of miR-99b-3p mimics (Fig. 6B), confirming that downregulation of CYLD and subsequent upregulation of RIPK1 are responsible for induction of apoptosis by the mimics. In contrast, introduction of miR-99b-3p mimics did not affect the viability of ME4405 and HEMn-MP cells (Fig. 6C), consistent with the finding of cell line-dependent regulation of CYLD expression by miR-99b-3p.
Figure 6.
RIPK1 rescues apoptosis induced by miR-99b-3p in Mel-CV and ME1007 cells. (A) Whole-cell lysates from Mel-CV and ME1007 cells transfected with indicated miRNA mimics were subjected to Western blotting. Data shown are representative of three individual experiments. (B) Mel-CV cells were transfected with indicated miRNA mimics, and plasmids were subjected to Western blotting (bottom) and PI/annexin V staining assays (top). Data shown are representative of three individual experiments (bottom) or means ± SEM (top). n = 3. **p < 0.01; ***p < 0.001, Student’s t-test. (C) ME4405 and HEMn-MP cells transfected with indicated miRNA mimics were subjected to CellTiter-Glo assays. Data shown are means ± SEM. n = 3. Student’s t-test.
DISCUSSION
As a tumor suppressor, CYLD inhibits cell proliferation in many cancer types including melanoma15. However, we have found in this study that the role of CYLD in regulating melanoma cell survival and proliferation varies considerably. While CYLD was necessary for survival of a proportion of melanoma cells, it inhibited cell proliferation in some others and did not impinge on cell viability in a third type of melanoma cells. Although these results are not entirely consistent with the well-established tumor-suppressive role of CYLD12,15,17,19, they reiterate the similarly well-documented heterogeneous features of melanoma9,10,42–44. For example, past studies have shown that the tumor suppressor p53 promotes melanoma cell survival upon endoplasmic reticulum (ER) stress and that inositol polyphosphate 4-phosphatase type II (INPP4B) that is a tumor suppressor in a number of cancer types functions as an oncogenic driver in a subset of melanomas45–47.
CYLD is known to inhibit canonical NF-κB activation through removing K63-linked polyubiquitin chains from a number of proteins involved in activation of the pathway such as TRAF2/6, Tak1, and NEMO and, thus, lead to their degradation by the proteasomal system48–51. Nevertheless, its inhibitory effect on melanoma cell proliferation is achieved through suppressing the noncanonical NF-κB pathway, resulting in reduced expression of cyclin D114,17. Moreover, regulation of JNK/AP-1 activation also plays a role in CYLD-mediated suppression of cyclin D1 expression19. Indeed, our results showed that the increase in ME4405 cell proliferation caused by CYLD knockdown was associated with upregulation of cyclin D1. On the other hand, we found that killing of melanoma cells in sensitive cell lines (Mel-CV and ME1007) by CYLD knockdown is due to induction of apoptosis, as it was associated with activation of the caspase cascade and was diminished by treatment with the general caspase inhibitor z-VAD-fmk.
The mechanism responsible for apoptosis induced by CYLD knockdown in sensitive melanoma cells (Mel-CV and ME1007) appeared to be upregulation of RIPK1. This was demonstrated by the findings that knockdown of CYLD resulted in upregulation of RIPK1 that was associated with enhancement in K63-linked polyubiquitination of the protein and that co-knockdown of RIPK1 diminished CYLD knockdown-induced apoptosis in sensitive cells. However, this seems inconsistent with our own recent results showing that RIPK1 is commonly increased in expression in melanoma cells and functions as an oncogenic driver through activation of NF-κB in melanoma21. Nevertheless, RIPK1 overexpression causes apoptosis through activation of the caspase cascade in many cell types22,52. Similarly, we have shown that overexpression of RIPK1 induces apoptosis in a proportion of melanoma cells21. We have proposed that the expression of RIP1 must be under tight control so that its cellular level remains constantly below a threshold to ensure avoidance of apoptosis21.
Although the molecular mechanism by which RIPK1 at high levels induces apoptosis in a selective population of melanoma cells remains unclear, it is conceivable that RIPK1 at high concentrations may trigger proximity-driven dimerization of caspase 8, thus leading to its activation and apoptosis, as the death domain (DD) of RIPK1 allows it to interact with other DD-containing proteins including FADD that can in turn recruit caspase 853–55. On the other hand, melanoma cells that can survive RIPK1 overexpression such as ME4405 and Mel-FH cells may possess intrinsic or acquired mechanisms that antagonize activation of caspase 8, most likely through NF-κB activation and subsequent upregulation of cFLIP56. Regardless, our results reveal a critical role of CYLD in controlling RIPK1 expression so that its cellular level remains constantly below a threshold to avoid induction of apoptosis.
Of note, the increased expression of RIPK1 upon CYLD knockdown did play a role in promotion of proliferation in ME4405 cells. Thus, along with previous findings that RIPK1 overexpression enhances proliferation of melanoma cells that have survived its apoptosis-inducing potential21,57, it suggests that other mechanisms such as increased NF-κB signaling resulting from enhanced activation of Bcl-3 are responsible for promotion of cell proliferation upon CYLD knockdown17. It remains unknown why CYLD does not impinge on survival and proliferation of an additional type of melanoma cells (Mel-FH). However, this was not due to CYLD mutations as all melanoma cell lines included in this study harbored wild-type CYLD. The different responses of melanoma cells to CYLD expression further highlight the biological heterogeneity of the disease.
Loss of function/expression of CYLD due to genetic mutations has been reported in various cancer types58–60. However, its expression in melanoma is primarily suppressed by transcriptional repression mediated by Snail1, a member of the Snail transcriptional repressors14,27. Moreover, a number of miRs including miR-767, miR-186, and miR-499-5p have been reported to be increased and play a role in suppression of CYLD expression in melanoma61–63. Our results showing that the CYLD protein is upregulated in a subset of melanoma cells whereas its transcript expression is, in general, lower in melanoma cells compared with melanocytes suggest that the increase in CYLD protein expression is caused by a posttranscriptional mechanism. The similar turnover rates of the CYLD protein among melanoma cell lines further pointed to the important role of translational regulation. Indeed, we identified loss of miR-99b-3p as a mechanism that selectively upregulated CYLD in Mel-CV and ME1007 but not in other melanoma cell lines. What determines the selectivity of miR-99b-3p in targeting CYLD mRNA remains to be clarified, but it is not due to variations in the 3′-UTR regions of CYLD as DNA sequencing of the region did not identify any mutations in the cell lines included in this study. It is well known that the expression and function of miRs are highly tissue and cell line dependent64,65. Although miR-767, miR-186, and miR-499-5p are known to target CYLD mRNA in melanoma, they are unlikely to be involved in CYLD upregulation in the subset of melanoma cells, as they were expressed at higher levels in melanoma cells compared with melanocytes61–63. Nevertheless, further investigations are needed to experimentally clarify whether these and others may compensate for the effect of miR-99b-3p on CYLD expression.
ACKNOWLEDGMENTS
This work was supported by the National Health and Medical Research Council (NHMRC) (X. D. Zhang: APP1147271 and L. Jin: APP1099947) and the Cancer Council NSW (X. D. Zhang: RG 16-12). L. Jin is a recipient of the Cancer Institute NSW Career Development Fellowship.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14(8):463–82. [DOI] [PubMed] [Google Scholar]
- 2. Gide TN, Quek C, Menzies AM, Tasker AT, Shang P, Holst J, Madore J, Lim SY, Velickovic R, Wongchenko M, Yan Y, Lo S, Carlino MS, Guminski A, Saw RPM, Pang A, McGuire HM, Palendira U, Thompson JF, Rizos H, Silva IPD, Batten M, Scolyer RA, Long GV, Wilmott JS. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell 2019;35(2):238–55.e6. [DOI] [PubMed] [Google Scholar]
- 3. Franke V, Berger DMS, Klop WMC, van der Hiel B, van de Wiel BA, Ter Meulen S, Wouters M, van Houdt WJ, van Akkooi ACJ. High response rates for T-VEC in early metastatic melanoma (stage IIIB/C-IVM1a). Int J Cancer 2019;145(4):974–8. [DOI] [PubMed] [Google Scholar]
- 4. Menzies AM, Long GV. Recent developments in melanoma therapy. JAMA Oncol. 2016;2(10):1259–60. [DOI] [PubMed] [Google Scholar]
- 5. Kozar I, Margue C, Rothengatter S, Haan C, Kreis S. Many ways to resistance: How melanoma cells evade targeted therapies. Biochim Biophys Acta Rev Cancer 2019;1871(2):313–22. [DOI] [PubMed] [Google Scholar]
- 6. Gide TN, Wilmott JS, Scolyer RA, Long GV. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin Cancer Res. 2018;24(6):1260–70. [DOI] [PubMed] [Google Scholar]
- 7. Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, Shannan B, Watters A, Perego M, Zhang G, Vultur A, Yin X, Liu Q, Anastopoulos IN, Wubbenhorst B, Wilson MA, Xu W, Karakousis G, Feldman M, Xu X, Amaravadi R, Gangadhar TC, Elder DE, Haydu LE, Wargo JA, Davies MA, Lu Y, Mills GB, Frederick DT, Barzily-Rokni M, Flaherty KT, Hoon DS, Guarino M, Bennett JJ, Ryan RW, Petrelli NJ, Shields CL, Terai M, Sato T, Aplin AE, Roesch A, Darr D, Angus S, Kumar R, Halilovic E, Caponigro G, Jeay S, Wuerthner J, Walter A, Ocker M, Boxer MB, Schuchter L, Nathanson KL, Herlyn M. A Comprehensive patient-derived xenograft collection representing the heterogeneity of melanoma. Cell Rep. 2017;21(7):1953–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hachey SJ, Boiko AD. Therapeutic implications of melanoma heterogeneity. Exp Dermatol. 2016;25(7):497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boshuizen J, Koopman LA, Krijgsman O, Shahrabi A, van den Heuvel EG, Ligtenberg MA, Vredevoogd DW, Kemper K, Kuilman T, Song JY, Pencheva N, Mortensen JT, Foppen MG, Rozeman EA, Blank CU, Janmaat ML, Satijn D, Breij ECW, Peeper DS, Parren PWHI. Cooperative targeting of melanoma heterogeneity with an AXL antibody–drug conjugate and BRAF/MEK inhibitors. Nat Med. 2018;24(2):203–12. [DOI] [PubMed] [Google Scholar]
- 10. Grzywa TM, Paskal W, Wlodarski PK. Intratumor and intertumor heterogeneity in melanoma. Transl Oncol. 2017;10(6):956–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Birkeland E, Zhang S, Poduval D, Geisler J, Nakken S, Vodak D, Meza-Zepeda LA, Hovig E, Myklebost O, Knappskog S, Lønning PE. Patterns of genomic evolution in advanced melanoma. Nat Commun. 2018;9(1):2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, Jones C, Hansen J, Blair E, Hofmann B, Siebert R, Turner G, Evans DG, Schrander-Stumpel C, Beemer FA, van Den Ouweland A, Halley D, Delpech B, Cleveland MG, Leigh I, Leisti J, Rasmussen S. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet. 2000;25(2):160–5. [DOI] [PubMed] [Google Scholar]
- 13. Massoumi R. CYLD: A deubiquitination enzyme with multiple roles in cancer. Future Oncol. 2011;7(2):285–97. [DOI] [PubMed] [Google Scholar]
- 14. Massoumi R, Kuphal S, Hellerbrand C, Haas B, Wild P, Spruss T, Pfeifer A, Fassler R, Bosserhoff AK. Down-regulation of CYLD expression by Snail promotes tumor progression in malignant melanoma. J Exp Med. 2009;206(1):221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mathis BJ, Lai Y, Qu C, Janicki JS, Cui T. CYLD-mediated signaling and diseases. Curr Drug Targets 2015;16(4):284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun SC. CYLD: A tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17(1):25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 2006;125(4):665–77. [DOI] [PubMed] [Google Scholar]
- 18. Jono H, Lim JH, Chen LF, Xu H, Trompouki E, Pan ZK, Mosialos G, Li JD. NF-kappaB is essential for induction of CYLD, the negative regulator of NF-kappaB: Evidence for a novel inducible autoregulatory feedback pathway. J Biol Chem. 2004;279(35):36171–4. [DOI] [PubMed] [Google Scholar]
- 19. Ke H, Augustine CK, Gandham VD, Jin JY, Tyler DS, Akiyama SK, Hall RP, Zhang JY. CYLD inhibits melanoma growth and progression through suppression of the JNK/AP-1 and beta1-integrin signaling pathways. J Invest Dermatol. 2013;133(1):221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang L, Du F, Wang X. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 2008;133(4):693–703. [DOI] [PubMed] [Google Scholar]
- 21. Liu XY, Lai F, Yan XG, Jiang CC, Guo ST, Wang CY, Croft A, Tseng HY, Wilmott JS, Scolyer RA, Jin L, Zhang XD. RIP1 kinase is an oncogenic driver in melanoma. Cancer Res. 2015;75(8):1736–48. [DOI] [PubMed] [Google Scholar]
- 22. Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011;43(3):432–48. [DOI] [PubMed] [Google Scholar]
- 23. La T, Liu GZ, Farrelly M, Cole N, Feng YC, Zhang YY, Sherwin SK, Yari H, Tabatabaee H, Yan XG, Guo ST, Liu T, Thorne RF, Jin L, Zhang XD. A p53-responsive microRNA network promotes cancer cell quiescence. Cancer Res. 2018;78(23):6666–79. [DOI] [PubMed] [Google Scholar]
- 24. Wang JY, Liu GZ, Wilmott JS, La T, Feng YC, Yari H, Yan XG, Thorne RF, Scolyer RA, Zhang XD, Jin L. Skp2-mediated stabilization of MTH1 promotes survival of melanoma cells upon oxidative stress. Cancer Res. 2017;77(22):6226. [DOI] [PubMed] [Google Scholar]
- 25. Nguyen T, Zhang XD, Hersey P. Relative resistance of fresh isolates of melanoma to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Clin Cancer Res. 2001;7(3 Suppl):966s–73s. [PubMed] [Google Scholar]
- 26. Ye Y, Jin L, Wilmott JS, Hu WL, Yosufi B, Thorne RF, Liu T, Rizos H, Yan XG, Dong L, Tay KH, Tseng HY, Guo ST, de Bock CE, Jiang CC, Wang CY, Wu M, Zhang LJ, Hersey P, Scolyer RA, Zhang XD. PI(4,5)P2 5-phosphatase A regulates PI3K/Akt signalling and has a tumour suppressive role in human melanoma. Nat Commun. 2013;4:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lei FX, Jin L, Liu XY, Lai F, Yan XG, Farrelly M, Guo ST, Zhao XH, Zhang XD. RIP1 protects melanoma cells from apoptosis induced by BRAF/MEK inhibitors. Cell Death Dis. 2018;9(6):679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhu H. Propidium iodide staining of cells for FACS analysis. Bio-protocol 2012;2(11):e195. [Google Scholar]
- 29. Ishikawa Y, Tsunoda K, Shibazaki M, Takahashi K, Akasaka T, Masuda T, Maesawa C. Downregulation of cylindromatosis gene, CYLD, confers a growth advantage on malignant melanoma cells while negatively regulating their migration activity. Int J Oncol. 2012;41(1):53–60. [DOI] [PubMed] [Google Scholar]
- 30. Wegner KW, Saleh D, Degterev A. Complex pathologic roles of RIPK1 and RIPK3: Moving beyond necroptosis. Trends Pharmacol Sci. 2017;38(3):202–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright A, Reiley WW, Chang M, Jin W, Lee AJ, Zhang M, Sun SC. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell 2007;13(5):705–16. [DOI] [PubMed] [Google Scholar]
- 32. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6(8):599–609. [DOI] [PubMed] [Google Scholar]
- 33. Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80. [DOI] [PubMed] [Google Scholar]
- 34. Zhang YY, Tabataba H, Liu XY, Wang JY, Yan XG, Farrelly M, Jiang CC, Guo ST, Liu T, Kao HY, Thorne RF, Zhang XD, Jin L. ACTN4 regulates the stability of RIPK1 in melanoma. Oncogene 2018;37(29):4033–45. [DOI] [PubMed] [Google Scholar]
- 35. Wang K, Liu F, Liu CY, An T, Zhang J, Zhou LY, Wang M, Dong YH, Li N, Gao JN, Zhao YF, Li PF. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ. 2016;23(8):1394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell 2008;30(6):689–700. [DOI] [PubMed] [Google Scholar]
- 37. Jaco I, Annibaldi A, Lalaoui N, Wilson R, Tenev T, Laurien L, Kim C, Jamal K, Wicky John S, Liccardi G, Chau D, Murphy JM, Brumatti G, Feltham R, Pasparakis M, Silke J, Meier P. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol Cell 2017;66(5):698–710.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dondelinger Y, Delanghe T, Priem D, Wynosky-Dolfi MA, Sorobetea D, Rojas-Rivera D, Giansanti P, Roelandt R, Gropengiesser J, Ruckdeschel K, Savvides SN, Heck AJR, Vandenabeele P, Brodsky IE, Bertrand MJM. Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat Commun. 2019;10(1):1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valinezhad Orang A, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014;2014:970607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 2012;3(3):311–30. [DOI] [PubMed] [Google Scholar]
- 41. Jiang CC, Croft A, Tseng HY, Guo ST, Jin L, Hersey P, Zhang XD. Repression of microRNA-768-3p by MEK/ERK signalling contributes to enhanced mRNA translation in human melanoma. Oncogene 2014;33(20):2577–88. [DOI] [PubMed] [Google Scholar]
- 42. Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jané-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352(6282):189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mesbah Ardakani N, Leslie C, Grieu-Iacopetta F, Lam WS, Budgeon C, Millward M, Amanuel B. Clinical and therapeutic implications of BRAF mutation heterogeneity in metastatic melanoma. Pigment Cell Melanoma Res. 2017;30(2):233–42. [DOI] [PubMed] [Google Scholar]
- 44. Reuben A, Spencer CN, Prieto PA, Gopalakrishnan V, Reddy SM, Miller JP, Mao X, De Macedo MP, Chen J, Song X, Jiang H, Chen PL, Beird HC, Garber HR, Roh W, Wani K, Chen E, Haymaker C, Forget MA, Little LD, Gumbs C, Thornton RL, Hudgens CW, Chen WS, Austin-Breneman J, Sloane RS, Nezi L, Cogdill AP, Bernatchez C, Roszik J, Hwu P, Woodman SE, Chin L, Tawbi H, Davies MA, Gershenwald JE, Amaria RN, Glitza IC, Diab A, Patel SP, Hu J, Lee JE, Grimm EA, Tetzlaff MT, Lazar AJ, Wistuba II, Clise-Dwyer K, Carter BW, Zhang J, Futreal PA, Sharma P, Allison JP, Cooper ZA, Wargo JA. Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom Med. 2017;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jiang CC, Lucas K, Avery-Kiejda KA, Wade M, deBock CE, Thorne RF, Allen J, Hersey P, Zhang XD. Up-regulation of Mcl-1 is critical for survival of human melanoma cells upon endoplasmic reticulum stress. Cancer Res. 2008;68(16):6708–17. [DOI] [PubMed] [Google Scholar]
- 46. Jin L, Hu WL, Jiang CC, Wang JX, Han CC, Chu P, Zhang LJ, Thorne RF, Wilmott J, Scolyer RA, Hersey P, Zhang XD, Wu M. MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc Natl Acad Sci USA 2011;108(38):15840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chi MN, Guo ST, Wilmott JS, Guo XY, Yan XG, Wang CY, Liu XY, Jin L, Tseng HY, Liu T, Croft A, Hondermarck H, Scolyer RA, Jiang CC, Zhang XD. INPP4B is upregulated and functions as an oncogenic driver through SGK3 in a subset of melanomas. Oncotarget 2015;6(37):39891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 2003;424(6950):801–5. [DOI] [PubMed] [Google Scholar]
- 49. Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 2003;424(6950):797–801. [DOI] [PubMed] [Google Scholar]
- 50. Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 2003;424(6950):793–6. [DOI] [PubMed] [Google Scholar]
- 51. Ahmed N, Zeng M, Sinha I, Polin L, Wei WZ, Rathinam C, Flavell R, Massoumi R, Venuprasad K. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat Immunol. 2011;12(12):1176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ikner A, Ashkenazi A. TWEAK induces apoptosis through a death-signaling complex comprising receptor-interacting protein 1 (RIP1), Fas-associated death domain (FADD), and caspase-8. J Biol Chem. 2011;286(24):21546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-kappaB signaling and cell death: So similar, yet so different. Cell Death Differ. 2017;24(7):1172–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He KL, Ting AT. A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol Cell Biol. 2002;22(17):6034–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 2009;137(4):721–35. [DOI] [PubMed] [Google Scholar]
- 56. Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR, Nachbur U, Rickard J, Anderton H, Moulin M, Lluis JM, Moujalled DM, Silke J, Vaux DL. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J Biol Chem. 2011;286(15):13282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jin L, Chen J, Liu XY, Jiang CC, Zhang XD. The double life of RIPK1. Mol Cell Oncol. 2016;3(1):e1035690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li YY, Chung GT, Lui VW, To KF, Ma BB, Chow C, Woo JK, Yip KY, Seo J, Hui EP, Mak MK, Rusan M, Chau NG, Or YY, Law MH, Law PP, Liu ZW, Ngan HL, Hau PM, Verhoeft KR, Poon PH, Yoo SK, Shin JY, Lee SD, Lun SW, Jia L, Chan AW, Chan JY, Lai PB, Fung CY, Hung ST, Wang L, Chang AM, Chiosea SI, Hedberg ML, Tsao SW, van Hasselt AC, Chan AT, Grandis JR, Hammerman PS, Lo KW. Exome and genome sequencing of nasopharynx cancer identifies NF-kappaB pathway activating mutations. Nat Commun. 2017;8:14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hajek M, Sewell A, Kaech S, Burtness B, Yarbrough WG, Issaeva N. TRAF3/CYLD mutations identify a distinct subset of human papillomavirus-associated head and neck squamous cell carcinoma. Cancer 2017;123(10):1778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nagy N, Farkas K, Kemeny L, Szell M. Phenotype–genotype correlations for clinical variants caused by CYLD mutations. Eur J Med Genet. 2015;58(5):271–8. [DOI] [PubMed] [Google Scholar]
- 61. Zhang K, Guo L. MiR-767 promoted cell proliferation in human melanoma by suppressing CYLD expression. Gene 2018;641:272–8. [DOI] [PubMed] [Google Scholar]
- 62. Qiu H, Yuan S, Lu X. miR-186 suppressed CYLD expression and promoted cell proliferation in human melanoma. Oncol Lett. 2016;12(4):2301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Long J, Pi X. lncRNA-MEG3 suppresses the proliferation and invasion of melanoma by regulating CYLD expression mediated by sponging miR-499-5p. Biomed Res Int. 2018;2018:2086564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E, Keller A. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fehlmann T, Ludwig N, Backes C, Meese E, Keller A. Distribution of microRNA biomarker candidates in solid tissues and body fluids. RNA Biol. 2016;13(11):1084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]