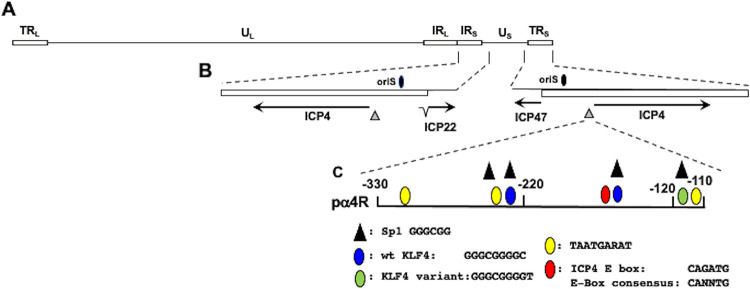
FIG 1.
Location of ICP4 gene, ICP4 enhancer, adjacent genes, and oriS within HSV-1 genome. (A) The prototypic HSV-1 genomic structure is shown. The viral repeat regions are shown as open rectangles. TRL is the terminal long repeat. IRL is the internal (or inverted) long repeat. TRS is the terminal short repeat. IRS is the internal (or inverted) short repeat. The unique long (UL) and the unique short (US) regions are each represented by a solid line. (B) Location of ICP4 coding regions and flanking genes in IRS (ICP22) or TRS (ICP47). The thin lines in ICP22 denote the location of an intron. Location of the origin of replication (oriS; denoted by black oval) and ICP4 enhancer sequences (−330 to −110; denoted by gray triangle). (C) Schematic of ICP4 enhancer fragment (pα4R) used in these studies. This fragment was inserted upstream of the minimal promoter of the firefly luciferase reporter plasmid, pGL4.24[luc2/minP]. Nucleotide position is given relative to the ICP4 transcription initiation site. Positions of consensus transcription factor binding sites are shown, and the key for these sites is below the schematic.