Dengue virus infects millions of people living in tropical areas of the world. Dengue virus-induced diseases can range from mild to severe with death.
KEYWORDS: antineutrophil cytoplasmic antibodies, dengue, vaccine
ABSTRACT
Any potential dengue virus (DENV) vaccine needs to elicit protective immunity against strains from all four serotypes to avoid potential antibody-dependent enhancement (ADE). In this study, four independent DENV envelope (E) glycoproteins were generated using wild-type E sequences from viruses isolated between 1943 and 2006 using computationally optimized broadly reactive antigen (COBRA) methodology. COBRA and wild-type E antigens were expressed on the surface of subvirion viral particles (SVPs). Four separate wild-type E antigens were used for each serotype. Mice vaccinated with wild-type DENV SVPs had anti-E IgG antibodies that neutralized serotype-specific viruses. COBRA DENV SVPs elicited a broader breadth of antibodies that neutralized strains across all four serotypes. Two COBRA DENV vaccine candidates that elicited the broadest breadth of neutralizing antibodies in mice were used to vaccinate rhesus macaques (Macaca mulatta) that either were immunologically naive to any DENV serotype or had preexisting antibodies to DENV. Antibodies elicited by COBRA DENV E immunogens neutralized all 12 strains of DENV in vitro, which was comparable to antibodies elicited by a tetravalent wild-type E SVP vaccination mixture. Therefore, using a single DENV COBRA E protein can elicit neutralizing antibodies against strains representing all four serotypes of DENV in both naive and dengue virus-preimmune populations.
IMPORTANCE Dengue virus infects millions of people living in tropical areas of the world. Dengue virus-induced diseases can range from mild to severe with death. An effective vaccine will need to neutralize viruses from all four serotypes of dengue virus without inducing enhanced disease. A dengue virus E vaccine candidate generated by computationally optimized broadly reactive antigen algorithms elicits broadly neutralizing protection for currently circulating strains from all four serotypes regardless of immune status. Most dengue vaccines in development formulate four separate components based on prM-E from a wild-type strain representing each serotype. Designing a monovalent vaccine that elicits protective immunity against all four serotypes is an effective and economical strategy.
INTRODUCTION
Dengue virus (DENV) is the most prevalent arbovirus worldwide and is endemic in more than 100 countries (1). Over half of the global population is at risk for DENV infection, with 100 million symptomatic cases being reported every year (2). It is estimated that there are 400 million infections a year, mainly in the tropical and subtropical regions of the Americas and Asia (3). There are four serotypes of the virus, DENV1 to -4 (4). They are genetically and antigenically distinct. Among the serotypes, ∼40% of the amino acids differ between the E glycoproteins, and there is a 9% difference within one serotype, resulting in different genotypes (5). In countries of endemicity, more than one serotype circulates (6). Cross-protective immunity is not long term, lasting about a year (7). Secondary infection with a heterologous serotype can lead to severe disease and mortality (8).
Dengue virus is a member of the Flavivirus family and is transmitted by Aedes aegypti and Aedes albopictus mosquitoes. It is enveloped and spherical, with a positive-sense single-stranded RNA genome that harbors one open reading frame (ORF) with 3 structural (capsid, matrix, and envelope) and 7 nonstructural (NS1, NS2a, NS2b, NS3, NS4a, NS4b, and NS5) proteins (9). The envelope glycoprotein (E) is the main target for neutralizing antibodies and is responsible for receptor binding and fusion (10). The membrane (M) and premembrane (prM) proteins are located on the surface envelope membrane of the virus. The immature prM form of the virus is usually found inside the host cell, where the prM protein acts as a chaperone for E protein folding in the endoplasmic reticulum (11). Cellular furin proteases cleave the prM and release the pr polypeptide. The remaining protein complex forms a mature, infectious virion that induces membrane fusion following an increase in the endosomal acidic pH (12, 13). Mature virions are released from the cell by exocytosis (14). In addition to full mature virions, noninfectious, immature subviral particles (SVPs) are also secreted from infected cells during viral replication (15). SVPs contain no genetic material and are produced by the expression of the prM-E gene cassette. These SVPs undergo the maturation process and acquire fusion abilities (16, 17).
Currently, the only licensed DENV vaccine, Dengvaxia, as well as two other leading candidates express DENV prM-E from either an attenuated, chimeric yellow fever virus (YFV) vector (18) or chimeric DENVs. The overall efficacy is ∼60%, and the vaccine is recommended for individuals between 9 and 45 years of age in countries of endemicity (19). Dengvaxia has been associated with vaccine enhancement in younger children and seronegative individuals (20–22). These vaccine candidates had low effectiveness during recent clinical trials, which may be due to the utilization of E gene inserts from wild-type (WT) viruses that did not match the currently circulating strains (23, 24).
To address the effects of genotype mismatch, novel DENV vaccines can be designed using multiple rounds of consensus building known as computationally optimized broadly reactive antigens (COBRAs). These result in DENV E immunogens that represent each DENV serotype spanning many genotypes (25). To generate COBRA sequences, 100 full-length E sequences from each DENV serotype, 1 to 4, from human infections were obtained from GenBank and used for COBRA consensus sequence generations. SVPs were produced from recombinant prM-E constructs. SVPs are noninfectious and a safer alternative to live-attenuated viral vaccines (26).
Two studies were performed to determine the immunogenicity and effectiveness of COBRA SVP vaccines: (i) a study in female C57BL/6 mice (aged 6 to 8 weeks) and (ii) a study in rhesus macaques (Macaca mulatta) that were immunologically naive to DENV or preimmune with antibodies to DENV serotype 1 or DENV serotype 2. Animals were vaccinated intramuscularly (i.m.) with DENV SVPs individually or in a tetravalent mixture. Immune sera were collected, total IgG antibody titers to DENV E were analyzed by an enzyme-linked immunosorbent assay (ELISA), and the ability to prevent virus infection in vitro was assessed in a 50% focus reduction neutralization test (FRNT50) against a panel of 12 prototype and modern strains from all four serotypes. Animals vaccinated with wild-type DENV SVPs expressed anti-E IgG antibodies that were specific for strains of each homologous serotype, and the elicited antibodies neutralized serotype-specific viruses. COBRA DENV SVPs elicited a broader breadth of antibodies that neutralized various strains across all four serotypes. One COBRA DENV E immunogen neutralized all 12 strains of DENV in vitro in both naive and preimmune groups, comparable to tetravalent SVP vaccination.
RESULTS
Dengue subviral particles.
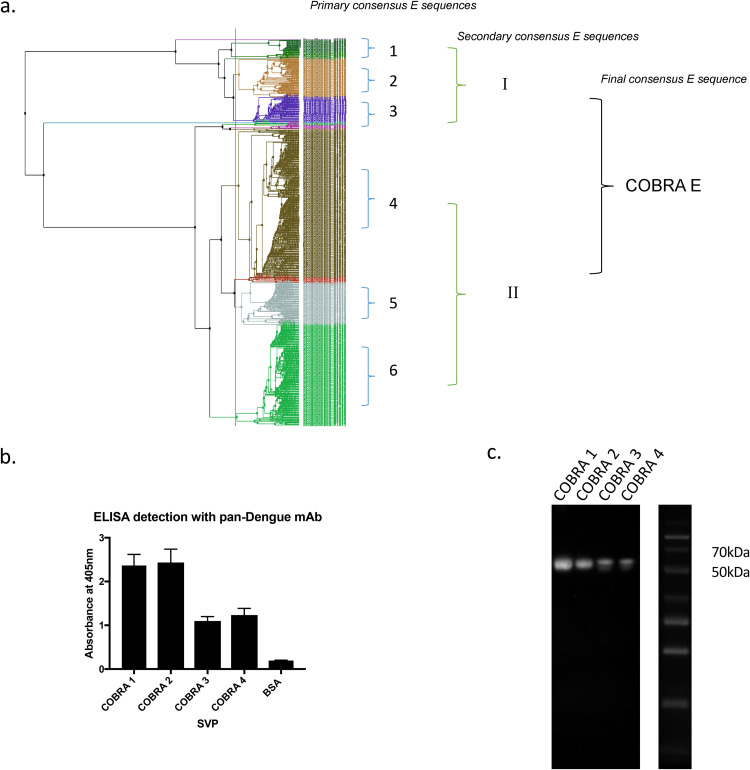
Using computationally optimized broadly reactive antigen (COBRA) technology, four unique dengue virus (DENV) envelope (E) genes were designed using ∼2,100 bp of full-length E sequences from human isolates (Fig. 1a). For each COBRA antigen, multiple rounds of sequences were aligned to create final COBRA E nucleotide sequences from 100 human clinical isolates downloaded from GenBank that span the many decades, from prototype to modern strains (1941 to 2006). Nucleotide sequences were translated into protein sequences using the standard genetic code. DENV E amino acid sequences were grouped by phylogenetic and geographic clustering. These clusters were aligned, and 6 primary consensus sequences were formed. Two secondary consensus sequences were formed from the primary consensus sequences. From these two sequences, the final COBRA E consensus sequence was generated.
FIG 1.
Design and construction of COBRA SVPs. (a) Dengue virus (DENV) E nucleotide sequences isolated from human infections from 1941 to 2006 were translated into protein sequences (100 from each serotype) and grouped by phylogenetic clustering and geographic location per serotype. Multiple rounds of sequence alignments were performed to retain common epitopes within the sequence. (b) ELISA to detect surface expression of DENV E using pan-dengue virus monoclonal antibody (mAb). Absorbance was measured at 405 nm. (c) Western blotting to detect E glycoprotein using panflavivirus monoclonal antibody. The protein ladder is on the right with relevant molecular weights indicated.
For the COBRA E sequence in Fig. 1a, primary cluster 1 isolates were mainly from Cambodia and Thailand, cluster 2 isolates were from Cambodia, and cluster 3 isolates were from Vietnam. The three primary consensus sequences from these three clusters subsequently formed an Asian-like secondary consensus sequence. Primary cluster 4 isolates were mainly from the United States and Venezuela, cluster 5 isolates were from Brazil, and cluster 6 isolates were from Nicaragua and Venezuela. The three primary consensus sequences from these three clusters subsequently formed an American-like secondary consensus sequence. The third and final consensus sequence yielded a COBRA E sequence.
Each COBRA E gene sequence was cloned into the vector pTR600 (27) and in frame with a premembrane (prM) sequence to express a subviral particle (SVP) following transfection into HEK 293T cells. Each SVP efficiently expressed each COBRA E protein on the particle surface (Fig. 1b) at the correct molecular weight, ∼55 kDa (Fig. 1c). COBRA SVPs had higher levels of COBRA 1 or COBRA 2 E proteins on their surface than SVPs expressing COBRA 3 or 4 E proteins.
COBRA SVP vaccinations in mice.
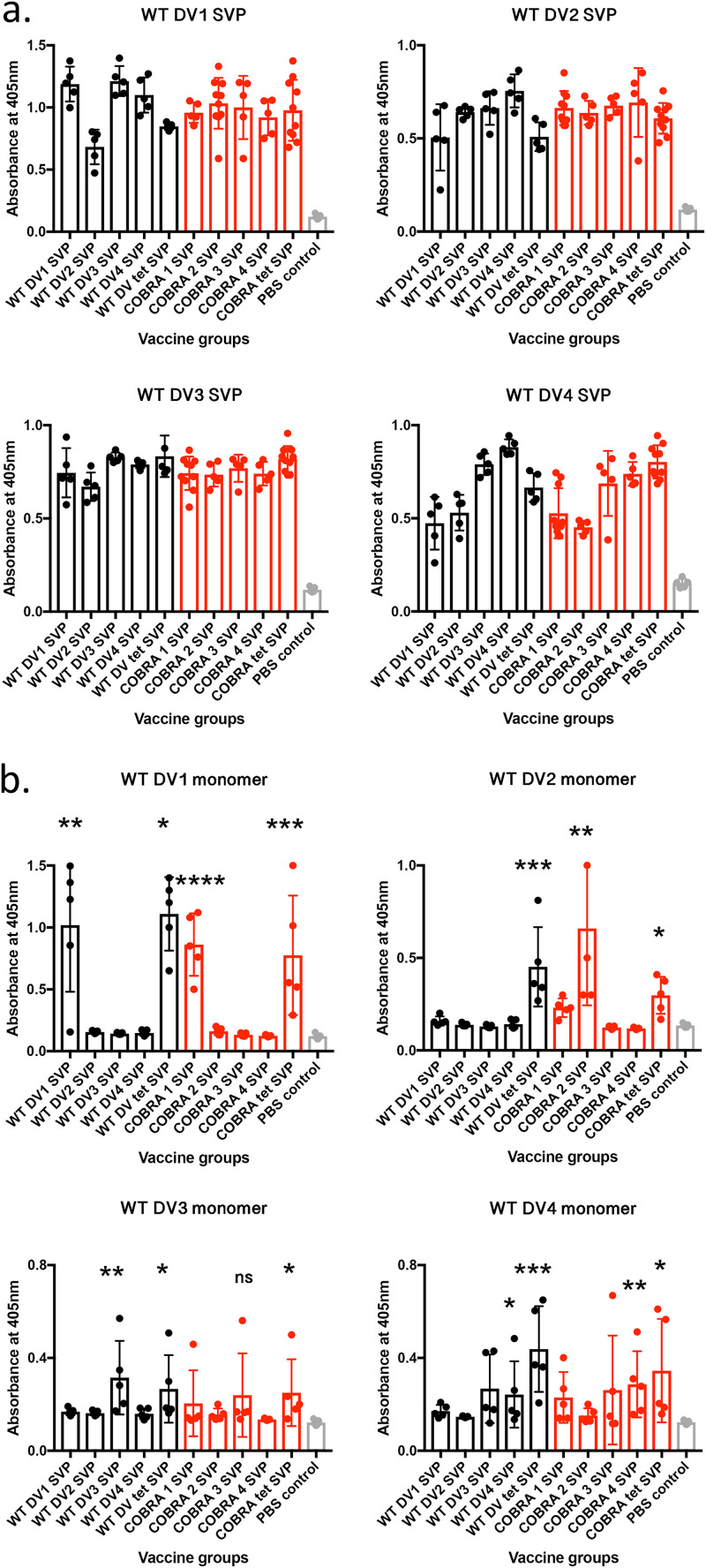
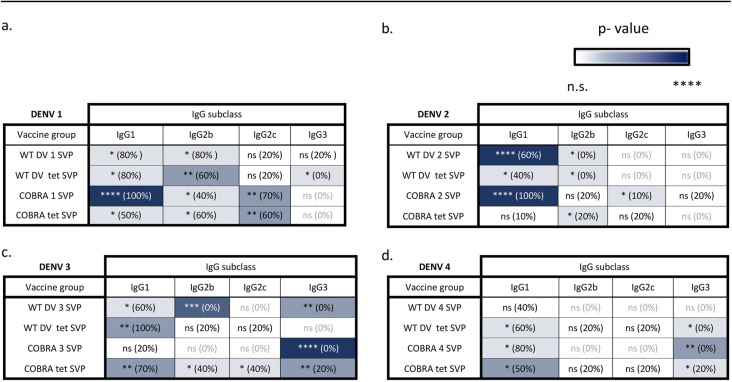
C57BL/6 mice were vaccinated with SVPs expressing the COBRA E or wild-type E dengue virus protein individually (monovalent) or in a tetravalent mixture. The vaccine consisted of 100 μg total SVP protein that was supplemented with Imject alum as an adjuvant. Four weeks after the final vaccination, sera were collected and tested for anti-E antibodies as well as for neutralization ability. All mice vaccinated with SVPs expressing WT or COBRA E antigens had significant cross-reactive IgG binding antibodies to any of the dengue virus serotypes expressed as SVPs (Fig. 2). One-way analyses of variance (ANOVAs) showed statistical significance between the vaccinated groups and the phosphate-buffered saline (PBS) control groups (see Table S1 in the supplemental material). However, each serum sample bound only to the commercially made soluble E protein that represented the serotype used for vaccination (homologous binding), with IgG (Fig. 2b). Mice vaccinated with SVP vaccines expressing COBRA 1 and 2 E antigens had high IgG1 titers to all four E antigens (Table 1). Additionally, COBRA 1 and 2 elicited IgG2c, which is associated with T helper 1 (Th1) responses.
FIG 2.
Seroconversion of mice given WT and COBRA SVP vaccinations. Each graph represents antibody binding to SVPs (a) or soluble E representing one of the four dengue virus serotypes (b). Vaccine groups are indicated on the x axis. Total IgG binding of sera (1:100 dilution) from individual mice was measured by the optical density (OD) values. The absorbance was measured at 405 nm. To determine the statistical significance of individual vaccine groups compared to the PBS control group, one-tailed unpaired t tests were done, with the confidence level at 95%. P values of <0.05 were considered statistically significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. DV1, dengue virus serotype 1; tet, tetravalent.
TABLE 1.
IgG subclasses elicited by WT and COBRA SVP vaccinesa
Each panel shows IgG subclass binding of homologous monovalent WT SVPs, WT tetravalent SVPs, COBRA E SVPs, and COBRA tetravalent SVPs to commercial E antigen representing DENV1 (a), DENV2 (b), DENV3 (c), and DENV4 (d). Asterisks represent statistical significance, measured against the PBS control group, using an unpaired t test with Welch’s correction, with the color intensity increasing with significance. ns, nonsignificant (P > 0.05); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
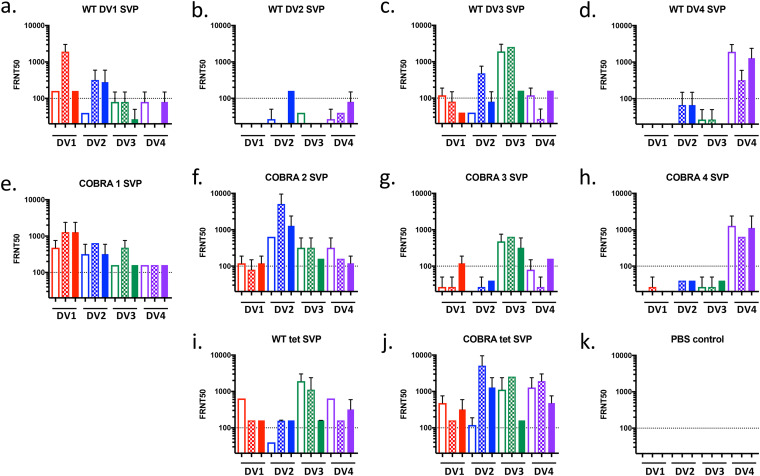
Next, focus reduction neutralization tests (FRNTs) were done to assess the breadth of the neutralization ability of the antibodies. Broadly neutralizing antibodies have been shown to be conformationally sensitive (28), which cannot be assessed by soluble-E-based assays. Sera were tested for the ability to neutralize a panel of 12 dengue viruses representing all 4 dengue virus serotypes. Modern strains (American and Asian) were chosen along with prototype strains since genotypes tend to separate by geographic location (29, 30) Mice vaccinated with each of the four SVPs expressing a WT E antigen elicited antibodies that neutralized primarily the viruses of the homologous serotype (Fig. 3a to d). In contrast, mice vaccinated with SVPs expressing the COBRA E antigens had antibodies that neutralized all four serotypes of dengue virus (Fig. 3e to h). COBRA 1 and COBRA 2 were comparable to tetravalent formulations (Fig. 3i and j). The COBRA tetravalent formulation had a more robust neutralizing antibody response than the WT tetravalent formulation. The COBRA 1, COBRA 2, and tetravalent formulations maintained broadly neutralizing titers in mice 20 weeks after the final vaccination (Fig. S1).
FIG 3.
Neutralization ability in WT and COBRA SVP vaccine groups. A focus reduction neutralization test (FRNT50) was performed on pooled sera from C57BL/6 mice vaccinated with monovalent WT SVPs (a to d), monovalent COBRA 1 to 4 SVPs (e to h), tetravalent SVPs (i and j), or the PBS control (k) against a panel of prototype (unfilled bars), American (checkered bars), and Asian (filled bars) strains from each serotype. Dashed lines indicate a 1:100 titer.
COBRA SVP vaccinations in naive and preimmune nonhuman primates.
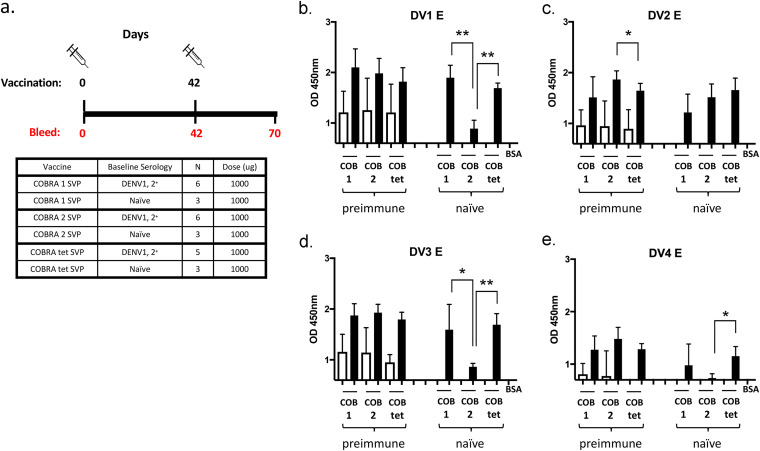
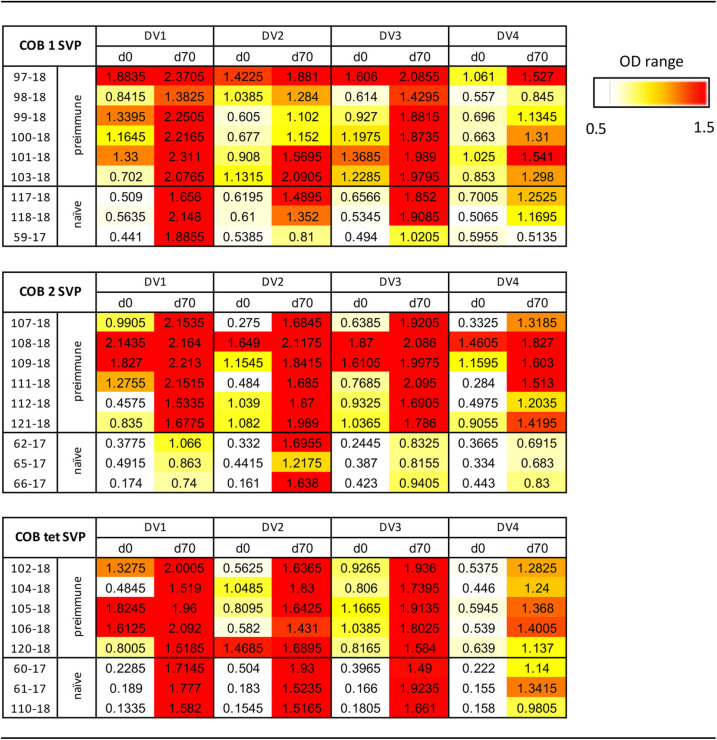
To determine the effectiveness of these vaccine candidates in preimmune populations, the top two monovalent COBRA E SVP vaccines were tested in rhesus macaques (Macaca mulatta) that were antibody positive for Brazilian strains of the DENV1 or DENV2 serotype (Fig. 4a). All preimmune monkeys had cross-reactive anti-E IgG antibodies both prior to and 4 weeks after vaccination (Fig. 4b to e and Table 2). Prior to vaccination, immunologically naive monkeys had little to no anti-IgG E antibodies to any of the DENV strains used in this study. However, after the first vaccination, all monkeys had anti-E antibodies (Fig. S2). After the second vaccination, all monkeys had significantly increased anti-E antibodies (Table S2). Comparing IgG binding to different dengue virus E proteins, there was no significant difference in the preimmune groups, except for COBRA 2 titers being significantly higher than with the COBRA tetravalent formulation to dengue virus serotype 2 (Fig. 4c). In naive groups, binding titers to COBRA 1 were comparable to those with the COBRA tetravalent formulation, while they were significantly higher than those with COBRA 2 in binding to E from serotypes 1 and 3. COBRA tetravalent binding was also higher than that of COBRA 2 to serotypes 1, 3, and 4 (Fig. 4).
FIG 4.
Seroconversion of NHPs given COBRA SVP vaccinations. (a) Schematic of COBRA SVP vaccinations in naive and preimmune NHPs. (b to e) Antibody binding to E of DENV1 to -4. Vaccine groups are listed on the x axis. Total IgG binding of sera (1:100 dilution) from individual NHPs was measured by the optical density (OD) values. Absorbance was measured at 405 nm. Unfilled bars, day 0; filled bars, day 70. Paired t tests were done to compare day 0 and day 70 values. Preimmune groups were analyzed by two-tailed paired t tests, and naive groups were analyzed by one-tailed paired t tests, with the confidence level at 95%. P values of <0.05 were considered statistically significant. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
TABLE 2.
Seroconversion of NHPs given COBRA SVP vaccinationa
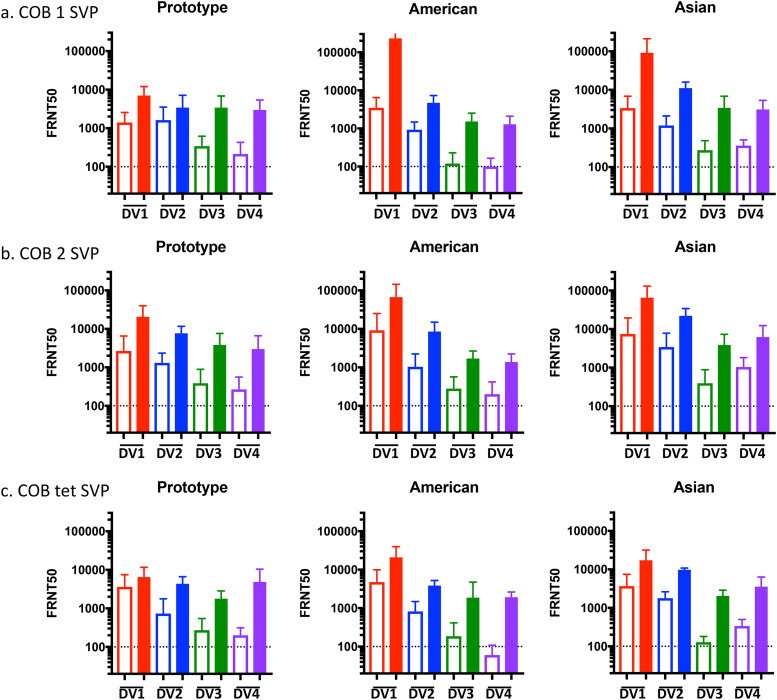
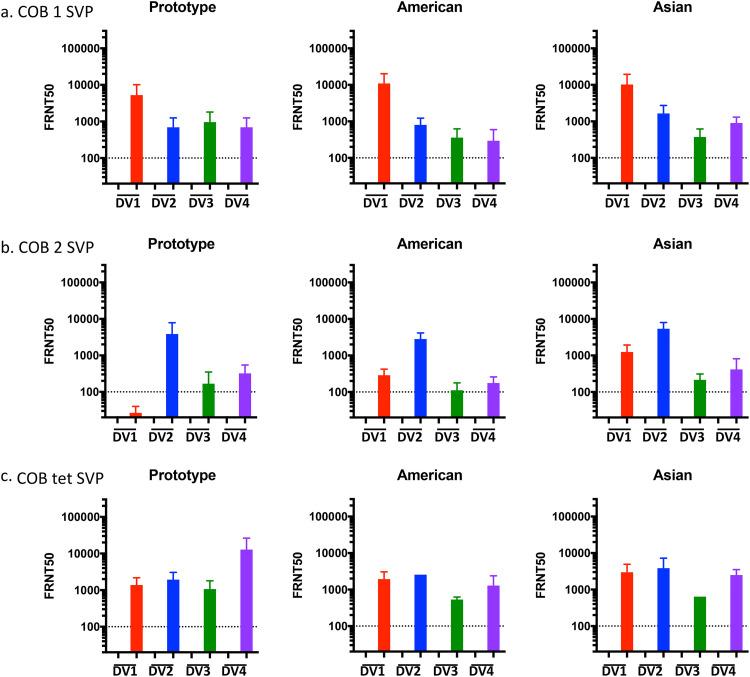
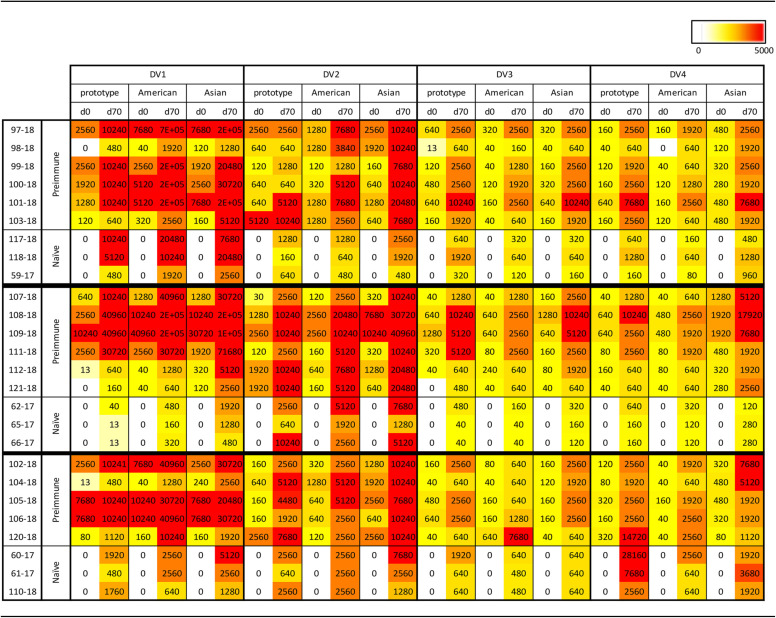
Monkeys vaccinated with COBRA 1 or 2 SVPs had neutralizing antibodies against all 11 to 12 DENV (Fig. 5 and 6 and Tables 3 and 4). Preimmune monkeys vaccinated with tetravalent COBRA SVPs elicited antibody titers statistically similar to those in animals vaccinated with either COBRA SVP vaccine (Table S3). Neutralizing titers (FRNT50s) were increased after vaccination in preimmune animals regardless of the vaccine administered (Fig. 5 and Table 3). In nonpreimmune monkeys, COBRA 1 SVP vaccines elicited neutralizing antibodies against all 12 strains, comparably to tetravalent COBRA, while COBRA 2 SVPs elicited neutralizing antibodies against 11 of the DENV strains (Fig. 6a to c). Neutralizing antibody titers elicited by COBRA 1 SVP vaccination were statistically similar to those elicited by COBRA tetravalent SVP vaccination, while COBRA 2 SVP vaccination elicited significantly lower neutralizing antibody titers than did COBRA tetravalent SVP vaccination (Table S3).
FIG 5.
Neutralization ability of COBRA SVP vaccines in preimmune NHPs. Shown are FRNT50 values for individual DENV-preimmune NHPs given COBRA 1 SVP (a), COBRA 2 SVP (b), and COBRA tetravalent SVP (c) vaccinations. Each graph represents a different genotype. DENV serotypes are indicated on the x axis. Dashed lines indicate a 1:100 titer. Unfilled bars, day 0; filled bars, day 70.
FIG 6.
Neutralization ability of COBRA SVP vaccines in naive NHPs. Shown are FRNT50 values of individual naive NHPs given COBRA 1 SVP (a), COBRA 2 SVP (b), and COBRA tetravalent SVP (c) vaccinations. Each graph represents a different genotype. DENV serotypes are indicated on the x axis. Dashed lines indicate a 1:100 titer. Unfilled bars, day 0; filled bars, day 70.
TABLE 3.
Neutralization ability of COBRA SVP vaccines in preimmune and naive NHPsa
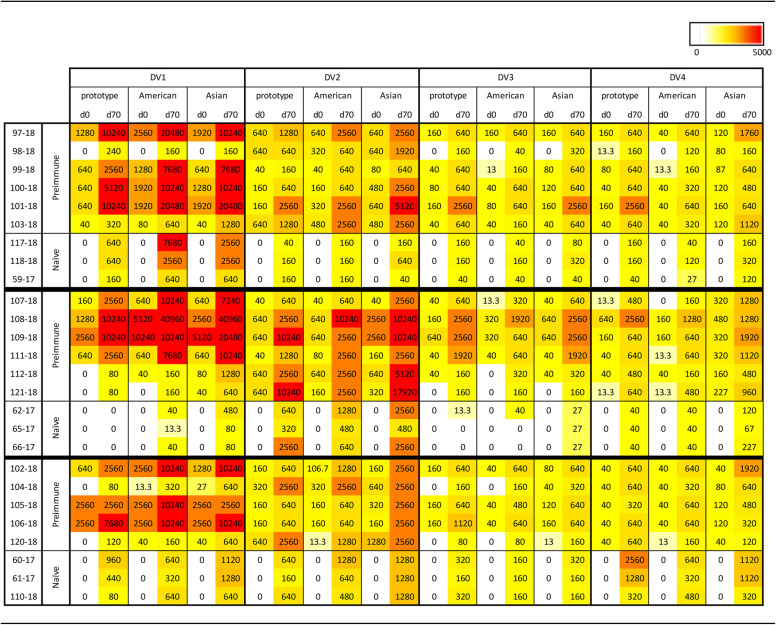
TABLE 4.
Neutralization ability of COBRA SVP vaccines in preimmune and naive NHPsa
Shown is a heat map of FRNT80 data.
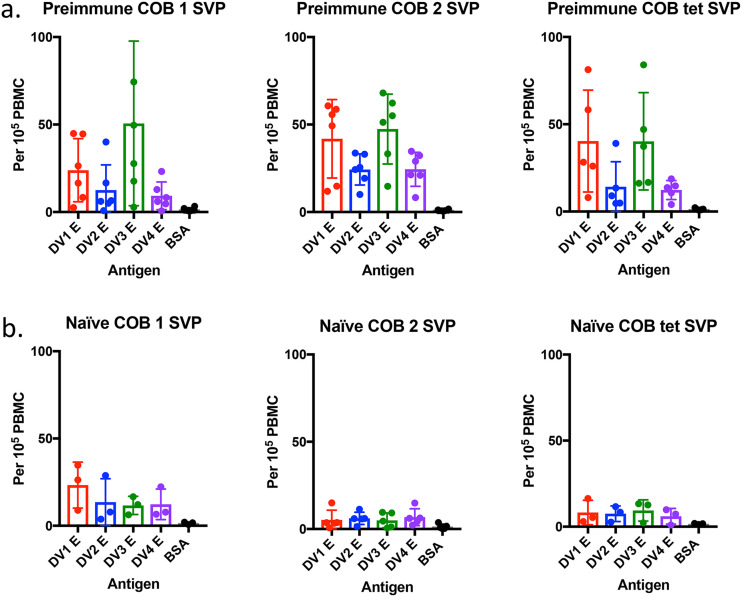
To study the memory response to dengue virus E, peripheral blood mononuclear cells (PBMCs) were collected at 4 weeks postboost and stimulated ex vivo with dengue virus full-length E. Antibody-secreting cells (ASCs) were enumerated by an enzyme-linked immunosorbent spot (ELISPOT) assay. Preimmune and naive monkeys vaccinated with COBRA vaccines had significantly higher dengue virus E-specific B cell responses than the nonspecific antigen control (Table S4). Preimmune monkeys had high numbers of ASCs against DENV1 and -3 (Fig. 7a). These same vaccines administered to naive animals had lower numbers of ASCs stimulated (Fig. 7b). Naive monkeys vaccinated with COBRA 1 SVPs elicited the highest number of ASCs, which correlated with the relatively high cross-reactive IgG and neutralizing antibody titers. Overall, COBRA 1 SVP vaccination in both preimmune and naive groups elicited memory B cells against all four serotypes comparable to the tetravalent formulation (Table S4). COBRA 2 may have elicited fewer cells, but each cell produced a higher number of neutralizing antibodies.
FIG 7.
B cell ELISPOT assay for DENV-specific antibody-secreting cells. NHP PBMCs were stimulated ex vivo with IL-2 and R848 for 5 days and then incubated with 5 μg/ml of DENV1 to -4 E proteins, anti-monkey IgG, or BSA. The total number of memory B cells (measured by their conversion to IgG-producing cells) was counted for each DENV E protein or BSA (x axis) in preimmune (a) and naive (b) NHP vaccination groups.
DISCUSSION
Making an effective vaccine for dengue virus has been challenging. The vaccine needs to be safe and protective against all four serotypes in populations with different serostatuses. With increased global travel and the spread of mosquitoes due to climate change, the risk of dengue virus infection continues to spread worldwide, while new strains are introduced to countries of endemicity (6). The COBRA methodology takes into account strains from various geographic locations and time periods in order to create a broadly protective immunogen. This technology has been successfully used to generate broadly reactive influenza virus vaccines (27, 31).
COBRA E sequences were designed from multiple consensus alignments of hundreds of human clinical isolates. The consensus alignments that we made were from clusters based on phylogeny and geographic region. It has been shown that strains from different geographic regions are more virulent than others of the same serotype (32). Increased global travel has allowed more virulent strains to spread and cause pandemics, displacing the previously circulating strain (33). Furthermore, the major circulating serotype often changes in countries of endemicity (6).
The COBRA E sequences were cloned into plasmid vectors in frame with prototype prM to be expressed on the surface of DENV SVPs after transient transfection. SVPs are more effective than recombinant E proteins as vaccine candidates since they still maintain fusogenic properties of live virus to activate endogenous as well as exogenous antigen recognition pathways (34). This process resulted in the COBRA 1 SVP vaccine, a promising universal DENV vaccine candidate that elicits neutralizing antibodies against all four serotypes in both naive and preimmune populations.
Correlates of protection against dengue virus are still not clearly defined, which contributes to the difficulties in vaccine design and assessment (24). Nonetheless, it is widely accepted that the humoral immune response, particularly neutralizing antibodies, is important for protecting against viral infection and preventing dengue virus-induced disease (35). The COBRA E antigen may elicit cross-neutralizing antibodies that target the quaternary structure of the E protein. COBRA SVP vaccinations elicited cross-reactive binding antibodies to both virus and virus-like particles expressing E proteins but not to soluble monomer versions of E (Fig. 2). Other studies show that monoclonal antibodies (mAbs) that bind to quaternary structures are effective at eliciting cross-neutralizing antibodies (28, 36, 37).
COBRA E antigens are an innovative approach for a universal DENV vaccine. Current dengue vaccine strategies need to formulate four separate components based on prM-E from wild-type strains of dengue virus that represent each dengue virus serotype. One of the challenges with this approach is the difficultly in achieving a balanced tetravalent formulation that may produce a skewed antibody response to one or more of the four dengue virus serotypes. In phase I clinical trials, DENV3 and DENV4 of the Dengvaxia vaccine replicated more efficiently than DENV1 or DENV2, which was later correlated with eliciting higher neutralizing antibody titers for DENV3 and DENV4 than for DENV1 or DENV2 in phase III trials (38). In contrast to vaccines that utilize four vaccine components, the single COBRA E vaccine will be more cost-effective to produce since it elicits cross-serotype antibodies similar to those of a vaccine containing four separate dengue virus components. Another advantage of using the dengue virus COBRA E-based vaccine is that it can be used with many different types of delivery platforms, such as RNA (39), DNA, expression from viral vectors, expression as a recombinant protein, as well as live-attenuated viruses.
One area of concern for any dengue vaccine is antibody-dependent enhancement (ADE). ADE occurs when preexisting antibodies to one DENV serotype do not neutralize but rather enhance a heterotypic infection (40, 41). Suboptimal levels of preexisting antibodies against dengue virus have been associated with severe DENV disease compared to no antibodies or high antibody levels (42). ADE usually occurs 2 years after a primary infection, and any cross-protective antibodies wane (43, 44). Thus, it is important for a DENV vaccine to elicit neutralizing antibodies against all four serotypes and maintain the robustness of the breadth of neutralization.
The only licensed vaccine, Dengvaxia, caused debate over its safety and efficacy due to vaccine enhancement in younger children and seronegative individuals (45). One theory for the low efficacy of these vaccines during recent clinical trials is that they utilized E gene inserts from wild-type viruses that did not match the currently circulating dengue virus strains. The COBRA immunogen design includes epitopes from modern strains, so it appears more comprehensive and relevant than vaccines using E antigens from older wild-type strains used in leading vaccine candidates.
Dengvaxia reduced the risk of symptomatic disease by 60% during the first 2 years after the vaccine was introduced (38). However, after the third year, preliminary data indicated that there was a higher risk of hospitalization. This trial will continue for another 6 years (45). COBRA SVP vaccination elicited memory B cells in nonhuman primates (NHPs) that were dengue virus specific (Fig. 7). In mice, the longevity of neutralizing antibodies was maintained 20 weeks after vaccination, although there was a significant decrease against 2 of the 12 strains in our panel (see Fig. S1 in the supplemental material).
The COBRA E SVP vaccine is a safer alternative to live viral vaccines while still maintaining the fusogenic properties of mature virions (16). The COBRA E vaccine also incorporates more relevant and comprehensive epitopes from strains representing multiple serotypes of dengue virus than the other leading dengue vaccine candidates (46, 47). The Dengvaxia vaccine uses Asian strains that were isolated between 1978 and 1988. DENVax uses Asian strains from 1964 to 1976, and TV003 uses strains isolated from 1974 to 1978.
COBRA 1 SVP vaccines elicited robust neutralizing antibodies (>1:200) against multiple dengue viral strains representing all four serotypes in both preimmune and nonpreimmune rhesus macaques. Although exact correlates of protection for neutralizing antibodies to dengue viruses are unknown, when testing nonreplicating DENV vaccines in rhesus macaques, neutralizing titers are recommended to be 1:200 (48). In a human cohort study of DENV infections, neutralization test titers of <1:100 were associated with severe disease (49). Due to the lack of an ideal animal model for DENV, clinical trials in humans to determine the safety and efficacy of the COBRA vaccine will need to be performed.
MATERIALS AND METHODS
Plasmid construction.
The full-length premembrane/envelope (prM/E) gene segments were cloned into the pTR600 expression plasmid. Dengue virus (DENV) E nucleotide sequences isolated from human infections from 1941 to 2006 were downloaded from the GenBank database (www.ncbi.nlm.nih.gov/genbank), and the sequences were translated into protein sequences (100 from each serotype). DENV E amino acid sequences were grouped by phylogenetic clustering and geographic location per serotype. Multiple rounds of sequence alignments were performed to retain common epitopes within the sequence. This methodology is referred to as a computationally optimized broadly reactive antigen (COBRA) design and has been described previously (27, 31). The final gene sequences were aligned using Align X (Vector NTI). The final amino acid COBRA E sequences as well as the wild-type (WT) E sequences from prototype wild-type DENV were reverse translated and optimized for expression in mammalian cells, including codon usage and RNA optimization (Gene Art, Regensburg, Germany). Prototype virus E sequences from the Hawaii (DENV1), NGC (DENV2), H87 (DENV3), and H241 (DENV4) strains (GenBank accession numbers X76219, M29095, M93130, and S66064) were optimized. WT and COBRA DENV E sequences representing all four subtypes of dengue virus were constructed. Plasmids were amplified using a Qiagen plasmid maxi kit, according to the manufacturer’s protocol (Qiagen, Germantown, MD, USA). Plasmids were verified using restriction enzyme digestions and examination of specific DNA fragment patterns following resolution by electrophoresis on a 1% agarose gel.
Transient transfection and purification of subviral particles.
Human embryonic kidney HEK 293T cells were transfected with 20 μg of each DNA plasmid using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). After incubation for 4 days at 37°C, the supernatants were collected, and cell debris was removed by centrifugation and vacuum filtration through a 0.22-μm sterile filter. Subviral particles (SVPs) were collected via ultracentrifugation (100,000 × g through 20% [wt/vol] glycerol) for 4 h at 4°C. The concentrated SVP pellets were subsequently resuspended in phosphate-buffered saline (PBS) and stored at −80°C. The protein concentration was determined by using the MicroBCA protein assay reagent kit (Thermo Fisher Scientific).
Verification of surface expression of DENV E.
For ELISAs, Nunc MaxiSorp plates (Thermo Fisher Scientific) were coated overnight at 4°C with SVPs diluted in PBS (1 μg/ml). Plates were washed with PBS containing 0.05% Tween (PBS-T) and blocked with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature (RT). Subsequently, pan-dengue virus monoclonal antibody (Acris antibody, catalog number AM01108PU-N; OriGene Technologies, Inc., Rockville, MD, USA) was added to the plates for 2 h. After washing 3 times, peroxidase-conjugated anti-mouse IgG antibody was added for 1 h before developing the plates using a pNpp substrate (SeraCare, Milford, MA, USA). The optical density (OD) was determined at 405 nm. For Western blotting, 1 μg of purified SVPs was added to a Bolt 4 to 12% Bis-Tris gel (Thermo Fisher Scientific) for SDS-PAGE under nonreducing conditions and electroblotted onto a polyvinylidene difluoride (PVDF) membrane. The samples were probed with panflavivirus mAb (5E284; Santa Cruz Biotechnology, Dallas, TX, USA) using the iBind Western device (Thermo Fisher Scientific). For the secondary antibody, goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (Southern Biotech, Birmingham, AL, USA) was used. The Clarity Western ECL substrate (Bio-Rad, Hercules, CA, USA) was used for chemiluminescence imaging.
Mice.
Female C57BL/6 mice (6 to 8 weeks of age; n = 5 to 10/group) were administered SVPs expressing WT or COBRA E antigens by intramuscular (i.m.) injection at weeks 0, 4, and 16 postvaccination. Each mouse received 100 μg of SVPs per vaccination with a 1:1 ratio per volume (25 μl/25 μl) of Imject alum adjuvant (Thermo Fisher Scientific). Mock-vaccinated mice were administered PBS. Blood was collected on weeks 4, 8, and 20 postvaccination and then centrifuged at 6,000 rpm for 10 min to separate the serum. Sera were frozen at −80°C.
Nonhuman primates.
Immunologically naive and preimmune rhesus macaques (Macaca mulatta) were vaccinated with monovalent or tetravalent DENV COBRA SVPs (1,000 μg per vaccination with Imject adjuvant) twice at 6-week intervals. Blood was collected at weeks 6 and 10, and sera and peripheral blood mononuclear cells (PBMCs) were isolated using BD Vacutainer cell preparation tubes (CPTs) with sodium heparin (Becton, Dickinson, Franklin Lakes, NJ, USA) according to the manufacturer’s protocol. Sera were frozen at −80°C. PBMCs were incubated in RPMI 1640 medium (Millipore Sigma, Burlington, MA, USA) containing 10% fetal bovine serum (FBS), 55 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/ml penicillin,100 μg/ml streptomycin, 10 mM HEPES, 1 mM sodium pyruvate, and 1% minimal essential medium (MEM) nonessential amino acids (all from Thermo Fisher Scientific).
Detection of anti-DENV E antibodies.
A quantitative ELISA was performed to detect anti-SVP or anti-E antibodies elicited by vaccination. Nunc MaxiSorp plates were coated (2 μg/ml) overnight at 4°C with either subviral particles expressing one of the wild-type DENV E proteins or soluble DENV E (MyBioSource, Inc., San Diego, CA, USA). Plates were washed with PBS-T and blocked with 1% BSA in PBS for 1 h at RT. Subsequently, a 1:100 dilution of serum samples was added to the plates for 2 h. Following three washes with PBS-T, peroxidase-conjugated anti-mouse IgG-Fc antibody (Bethyl Laboratories, Montgomery, TX, USA) or HRP-conjugated anti-monkey IgG(H+L) antibody (Thermo Fisher Scientific) was added for 1 h before the plates were developed using a pNpp or TMB (3,3′,5,5′-tetramethylbenzidine) substrate, respectively. The OD values were read at 405 nm for pNpp and 450 nm for TMB.
Dengue viruses.
Viruses used in this study were DENV1 strains DV1/US/Hawaii/1944, DV1/Vietnam/BID-V1792/2007, and DV1/Costa Rica/BC89/1994; DENV2 strains DV2/New Guinea/NGC/1944, DV2/Vietnam/BID-V1007/2006, and DV2/US/BID-V594/2006; DENV3 strains DV3/Philippines/H87/1956, DV3/Sri Lanka/271242/1991, and DV3/US/BID-V1619/2005; and DENV4 strains DV4/Philippines/H241/1956, DV4/Malaysia/BC13/1997, and DV4/US/BC258/1994.
Neutralization assay.
Neutralization activities in collected antisera were determined using a focus reduction neutralization test (FRNT). Sera were heat inactivated at 56°C for 30 min and then serially diluted (4-fold). Sera were incubated with dengue viruses (100 focus-forming units [FFU]) representing each of the 4 serotypes for 60 min at 37°C. Three dengue viruses were used for each serotype that represented the prototype, modern Asian, and modern American strains. Each dengue virus was mixed with the serially diluted antisera and used to infect a single-cell monolayer of Vero cells that were plated in 96-well plates for 90 min at 37°C. Cells were overlaid with methylcellulose (1% [wt/vol]). Cells were incubated for 3 days at 37°C (2 days for DENV4 strains). Cells were washed with PBS and fixed with a mixture of acetone and methanol (foci were stained using a Vectastain ABC kit according to the manufacturer’s protocols [Vector Laboratories, Burlingame, CA, USA]). Anti-DENV E monoclonal antibodies were used to detect virally infected cells. For DENV1 and -3 strains, a pan-dengue virus antibody (catalog number AM01108PU-N; OriGene Technologies, Rockville, MD, USA) was used; for DENV2, the monoclonal antibody 9.F.10 (Santa Cruz Biotechnology, Dallas, TX, USA) was used; and for DENV4, the monoclonal antibody 4G2 (Millipore Sigma, Billerica, MA, USA) was used. Foci were imaged using the CTL ImmunoSpot analyzer (Cellular Technology Limited, Cleveland, OH, USA). The FRNT50 was determined by calculating the point at which sera reduced dengue virus foci by 50% compared to serum-free, dengue virus control wells.
B cell ELISPOT assay.
Nonhuman primate (NHP) PBMCs were stimulated ex vivo with interleukin-2 (IL-2) and R848 (Mabtech) for 5 days. A multiscreen HTS plate (Millipore Sigma) was prewetted with 35% ethanol and coated with 5 μg/ml of DENV1 to -4 peptides (MyBioSource), anti-monkey IgG (Mabtech), or BSA. PBMCs were added in serial dilutions starting at 300,000 cells/well in RPMI 1640 medium supplemented with 55 μM 2-mercaptoethanol, 2 mM l-glutamine, 100 U/ml penicillin,100 μg/ml streptomycin, 10 mM HEPES, 1 mM sodium pyruvate, and 1% MEM nonessential amino acids and incubated at 37°C for 24 h. The cells were washed five times with PBS, and anti-monkey IgG-biotin (Mabtech) was added overnight at 4°C. The following day, plates were washed five times with PBS. Streptavidin-HRP (Mabtech) was added, and the plates were incubated at room temperature for 2 h. After washing five times with PBS, spots were developed with a TMB solution (Vector Laboratories). Spots were imaged using a CTL ImmunoSpot analyzer (Cellular Technology Limited, Cleveland, OH, USA) and counted using ImmunoSpot software version 5.1.36 Professional SC.
Statistical analysis.
To determine the statistical significance of individual mouse vaccine groups compared to the PBS control group, one-tailed unpaired t tests were done, with a confidence level at 95%. Vaccine groups were compared for ELISA antibody binding against the PBS control group with soluble E from all four serotypes. Differences in murine IgG subclass responses were analyzed by an unpaired t test with Welch’s correction.
To determine statistically significant differences in NHP antibody binding before and after vaccination with COBRA SVPs, paired t tests were done to compare day 0 and day 70 ELISA and FRNT data for all 4 serotypes. Preimmune NHP groups were analyzed by two-tailed paired t tests, and naive NHP groups were analyzed by one-tailed paired t tests.
Statistical analyses were performed using Prism (GraphPad Software, La Jolla, CA, USA). P values of less than 0.05 were considered statistically significant (ns, not significant [P > 0.05]; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI. 2012. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis 6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzman MG, Harris E. 2015. Dengue. Lancet 385:453–465. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas SJ, Endy TP, Rothman AL. 2014. Flaviviruses: dengue, p 351–381. In Kaslow R, Stanberry LR, Le Duc JW (ed), Viral infections of humans: epidemiology and control, 5th ed. Springer, New York, NY. [Google Scholar]
- 5.Bennett SN. 2010. Evolutionary dynamics of dengue virus, p 157–172. In Hanley KA, Weaver SC (ed), Frontiers in dengue virus research. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 6.Messina JP, Brady OJ, Scott TW, Zou C, Pigott DM, Duda KA, Bhatt S, Katzelnick L, Howes RE, Battle KE, Simmons CP, Hay SI. 2014. Global spread of dengue virus types: mapping the 70 year history. Trends Microbiol 22:138–146. doi: 10.1016/j.tim.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reich NG, Shrestha S, King AA, Rohani P, Lessler J, Kalayanarooj S, Yoon I-K, Gibbons RV, Burke DS, Cummings DA. 2013. Interactions between serotypes of dengue highlight epidemiological impact of cross-immunity. J R Soc Interface 10:20130414. doi: 10.1098/rsif.2013.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libraty DH, Endy TP, Houng H-SH, Green S, Kalayanarooj S, Suntayakorn S, Chansiriwongs W, Vaughn DW, Nisalak A, Ennis FA, Rothman AL. 2002. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis 185:1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 9.Chambers TJ, Hahn CS, Galler R, Rice CM. 1990. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 10.Halstead S. 2008. Dengue. Tropical medicine: science and practice, vol 5. Imperial College Press, London, United Kingdom. [Google Scholar]
- 11.Courageot M-P, Frenkiel M-P, Dos Santos CD, Deubel V, Desprès P. 2000. α-Glucosidase inhibitors reduce dengue virus production by affecting the initial steps of virion morphogenesis in the endoplasmic reticulum. J Virol 74:564–572. doi: 10.1128/jvi.74.1.564-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 13.Yu I-M, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 14.Heinz F, Auer G, Stiasny K, Holzmann H, Mandl C, Guirakhoo F, Kunz C. 1994. The interactions of the flavivirus envelope proteins: implications for virus entry and release. Arch Virol Suppl 9:339–348. doi: 10.1007/978-3-7091-9326-6_34. [DOI] [PubMed] [Google Scholar]
- 15.van der Schaar HM, Rust MJ, Waarts B-L, van der Ende-Metselaar H, Kuhn RJ, Wilschut J, Zhuang X, Smit JM. 2007. Characterization of the early events in dengue virus cell entry by biochemical assays and single-virus tracking. J Virol 81:12019–12028. doi: 10.1128/JVI.00300-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell P, Brandt W, Dalrymple J. 1980. Chemical and antigenic structure of flaviviruses, p 503–529. In Schlesinger RW (ed), The togaviruses: biology, structure, replication. Academic Press, New York, NY. [Google Scholar]
- 17.Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell 7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- 18.Guy B, Briand O, Lang J, Saville M, Jackson N. 2015. Development of the Sanofi Pasteur tetravalent dengue vaccine: one more step forward. Vaccine 33:7100–7111. doi: 10.1016/j.vaccine.2015.09.108. [DOI] [PubMed] [Google Scholar]
- 19.Torresi J, Ebert G, Pellegrini M. 2017. Vaccines licensed and in clinical trials for the prevention of dengue. Hum Vaccin Immunother 13:1059–1072. doi: 10.1080/21645515.2016.1261770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halstead SB. 2016. Licensed dengue vaccine: public health conundrum and scientific challenge. Am J Trop Med Hyg 95:741–745. doi: 10.4269/ajtmh.16-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halstead SB. 2016. Critique of World Health Organization recommendation of a dengue vaccine. J Infect Dis 214:1793–1795. doi: 10.1093/infdis/jiw340. [DOI] [PubMed] [Google Scholar]
- 22.Aguiar M, Stollenwerk N, Halstead SB. 2016. The risks behind Dengvaxia recommendation. Lancet Infect Dis 16:882–883. doi: 10.1016/S1473-3099(16)30168-2. [DOI] [PubMed] [Google Scholar]
- 23.Sabchareon A, Wallace D, Sirivichayakul C, Limkittikul K, Chanthavanich P, Suvannadabba S, Jiwariyavej V, Dulyachai W, Pengsaa K, Wartel TA, Moureau A, Saville M, Bouckenooghe A, Viviani S, Tornieporth NG, Lang J. 2012. Protective efficacy of the recombinant, live-attenuated, CYD tetravalent dengue vaccine in Thai schoolchildren: a randomised, controlled phase 2b trial. Lancet 380:1559–1567. doi: 10.1016/S0140-6736(12)61428-7. [DOI] [PubMed] [Google Scholar]
- 24.de Silva AM, Harris E. 2018. Which dengue vaccine approach is the most promising, and should we be concerned about enhanced disease after vaccination? The path to a dengue vaccine: learning from human natural dengue infection studies and vaccine trials. Cold Spring Harb Perspect Biol 10:a029371. doi: 10.1101/cshperspect.a029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross T, Tang X, Lu H, Olagnier D, Kirchenbaum G, Evans J. 2014. COBRA-based dengue tetravalent vaccine elicits neutralizing antibodies against all four dengue serotypes. J Vaccine Immunotechnol 1:7. [Google Scholar]
- 26.Roldao A, Mellado MC, Castilho LR, Carrondo MJ, Alves PM. 2010. Virus-like particles in vaccine development. Expert Rev Vaccines 9:1149–1176. doi: 10.1586/erv.10.115. [DOI] [PubMed] [Google Scholar]
- 27.Giles BM, Ross TM. 2011. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 29:3043–3054. doi: 10.1016/j.vaccine.2011.01.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, Grimes JM, Tsai W-Y, Lai C-Y, Wang W-K, Malasit P, Farrar J, Simmons CP, Zhou ZH, Rey FA, Mongkolsapaya J, Screaton GR. 2015. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 16:170–177. doi: 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mir D, Romero H, de Carvalho LMF, Bello G. 2014. Spatiotemporal dynamics of DENV-2 Asian-American genotype lineages in the Americas. PLoS One 9:e98519. doi: 10.1371/journal.pone.0098519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes EC, Twiddy SS. 2003. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol 3:19–28. doi: 10.1016/S1567-1348(03)00004-2. [DOI] [PubMed] [Google Scholar]
- 31.Carter DM, Darby CA, Lefoley BC, Crevar CJ, Alefantis T, Oomen R, Anderson SF, Strugnell T, Cortés-Garcia G, Vogel TU, Parrington M, Kleanthous H, Ross TM. 2016. Design and characterization of a computationally optimized broadly reactive hemagglutinin vaccine for H1N1 influenza viruses. J Virol 90:4720–4734. doi: 10.1128/JVI.03152-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rico-Hesse R. 2010. Dengue virus virulence and transmission determinants. Curr Top Microbiol Immunol 338:45–55. doi: 10.1007/978-3-642-02215-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cologna R, Armstrong PM, Rico-Hesse R. 2005. Selection for virulent dengue viruses occurs in humans and mosquitoes. J Virol 79:853–859. doi: 10.1128/JVI.79.2.853-859.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noad R, Roy P. 2003. Virus-like particles as immunogens. Trends Microbiol 11:438–444. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- 35.Cedillo-Barrón L, García-Cordero J, Bustos-Arriaga J, León-Juárez M, Gutiérrez-Castañeda B. 2014. Antibody response to dengue virus. Microbes Infect 16:711–720. doi: 10.1016/j.micinf.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 36.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WMPB, White LJ, Diamond MS, Baric RS, Crowe JE, Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Natl Acad Sci U S A 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney M-C, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A, Girard-Blanc C, Petres S, Shepard WE, Desprès P, Arenzana-Seisdedos F, Dussart P, Mongkolsapaya J, Screaton GR, Rey FA. 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 520:109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 38.Hadinegoro SR, Arredondo-Garcia JL, Capeding MR, Deseda C, Chotpitayasunondh T, Dietze R, Muhammad Ismail HI, Reynales H, Limkittikul K, Rivera-Medina DM, Tran HN, Bouckenooghe A, Chansinghakul D, Cortes M, Fanouillere K, Forrat R, Frago C, Gailhardou S, Jackson N, Noriega F, Plennevaux E, Wartel TA, Zambrano B, Saville M, CYD-TDV Dengue Vaccine Working Group. 2015. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 373:1195–1206. doi: 10.1056/NEJMoa1506223. [DOI] [PubMed] [Google Scholar]
- 39.Sahin U, Karikó K, Türeci Ö. 2014. mRNA-based therapeutics—developing a new class of drugs. Nat Rev Drug Discov 13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 40.Halstead S, O’Rourke E. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med 146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halstead SB. 2007. Dengue. Lancet 370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 42.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E. 2017. Antibody-dependent enhancement of severe dengue disease in humans. Science 358:929–932. doi: 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montoya M, Gresh L, Mercado JC, Williams KL, Vargas MJ, Gutierrez G, Kuan G, Gordon A, Balmaseda A, Harris E. 2013. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 7:e2357. doi: 10.1371/journal.pntd.0002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang TK, Ghebremariam SL, Gresh L, Gordon A, Halloran ME, Katzelnick LC, Rojas DP, Kuan G, Balmaseda A, Sugimoto J, Harris E, Longini IM, Jr, Yang Y. 2019. Effects of infection history on dengue virus infection and pathogenicity. Nat Commun 10:1246. doi: 10.1038/s41467-019-09193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dans AL, Dans LF, Lansang MAD, Silvestre MAA, Guyatt GH. 2018. Controversy and debate on dengue vaccine series—paper 1. Review of a licensed dengue vaccine: inappropriate subgroup analyses and selective reporting may cause harm in mass vaccination programs. J Clin Epidemiol 95:137–139. doi: 10.1016/j.jclinepi.2017.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Osorio JE, Velez ID, Thomson C, Lopez L, Jimenez A, Haller AA, Silengo S, Scott J, Boroughs KL, Stovall JL, Luy BE, Arguello J, Beatty ME, Santangelo J, Gordon GS, Huang CYH, Stinchcomb DT. 2014. Safety and immunogenicity of a recombinant live attenuated tetravalent dengue vaccine (DENVax) in flavivirus-naive healthy adults in Colombia: a randomised, placebo-controlled, phase 1 study. Lancet Infect Dis 14:830–838. doi: 10.1016/S1473-3099(14)70811-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Precioso AR, Palacios R, Thomé B, Mondini G, Braga P, Kalil J. 2015. Clinical evaluation strategies for a live attenuated tetravalent dengue vaccine. Vaccine 33:7121–7125. doi: 10.1016/j.vaccine.2015.09.105. [DOI] [PubMed] [Google Scholar]
- 48.Hombach J, Cardosa MJ, Sabchareon A, Vaughn DW, Barrett AD. 2007. Scientific consultation on immunological correlates of protection induced by dengue vaccines: report from a meeting held at the World Health Organization 17–18 November 2005. Vaccine 25:4130–4139. doi: 10.1016/j.vaccine.2007.02.079. [DOI] [PubMed] [Google Scholar]
- 49.Salje H, Cummings DAT, Rodriguez-Barraquer I, Katzelnick LC, Lessler J, Klungthong C, Thaisomboonsuk B, Nisalak A, Weg A, Ellison D, Macareo L, Yoon I-K, Jarman R, Thomas S, Rothman AL, Endy T, Cauchemez S. 2018. Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature 557:719–723. doi: 10.1038/s41586-018-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.