Fig. 1.
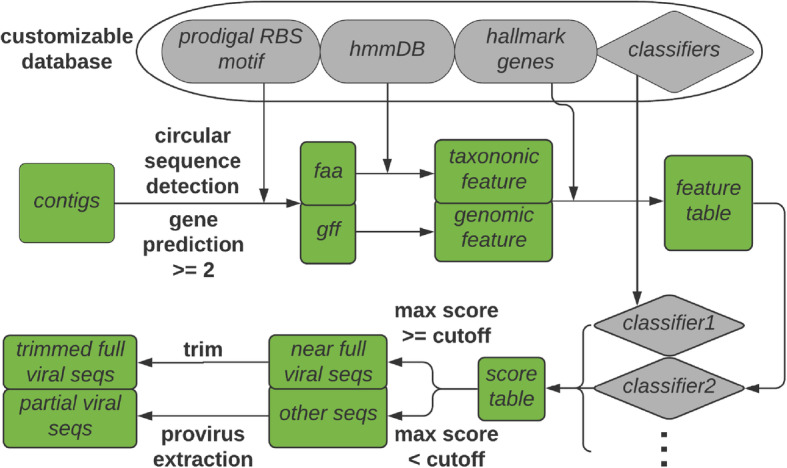
Overview of the VirSorter2 framework. Schematic of the viral prediction pipeline used in VirSorter2. “hmmDB” represents databases of HMM profiles including viral HMMs from Xfam (described in the “Methods” section) and viral protein families (VPF) from JGI Earth’s Virome [17], and cellular HMMs (archaeal, bacterial, eukaryotic) as well as “mixed” HMMs (not specific to either virus or cellular organisms) from Pfam [43]. A default cutoff of 30 is used for the HMM searches. “Classifiers” refers to random forest classifiers trained on known viral genomes and cellular genomes from different viral groups (see “Training classifiers” section in “Methods”). The default max score cutoff is set to 0.5
