Fig. 3.
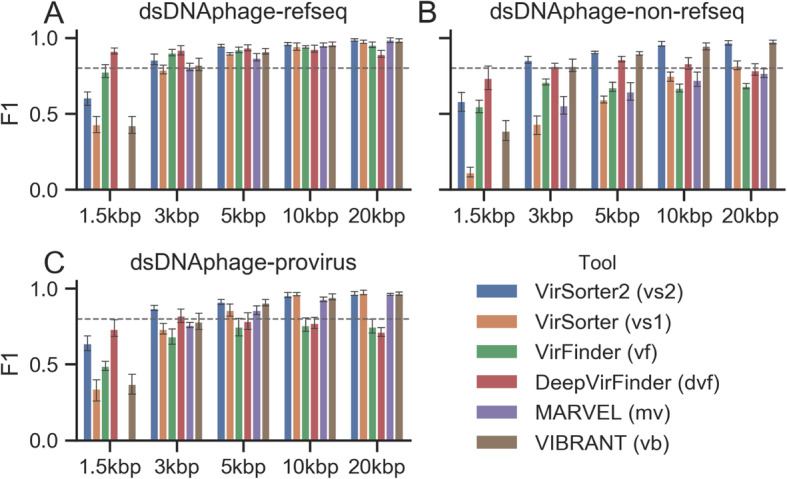
Tool performances on dsDNA phages from different data sources. VirSorter2 consistently has comparable or better performance than existing tools in identifying dsDNA phages. Genome fragments of different lengths (x-axis) are generated from genomes in the order Caudovirales in NCBI Viral RefSeq (a), proviruses extracted from microbial genomes in NCBI RefSeq (b) [48], and other sources (c) [15, 16]. An equal number (50) of viral and non-viral (archaea and bacteria, fungi and protozoa, and plasmids) genome fragments were combined as an input for the tested tools. Error bars show 95% confidence intervals over five replicates (100 sequences each as described above). F1 score is used as the metric (y-axis) to compare tools, while detailed recall and precision results are available in Figs. S1 and S2. The dotted line is y = 0.8
