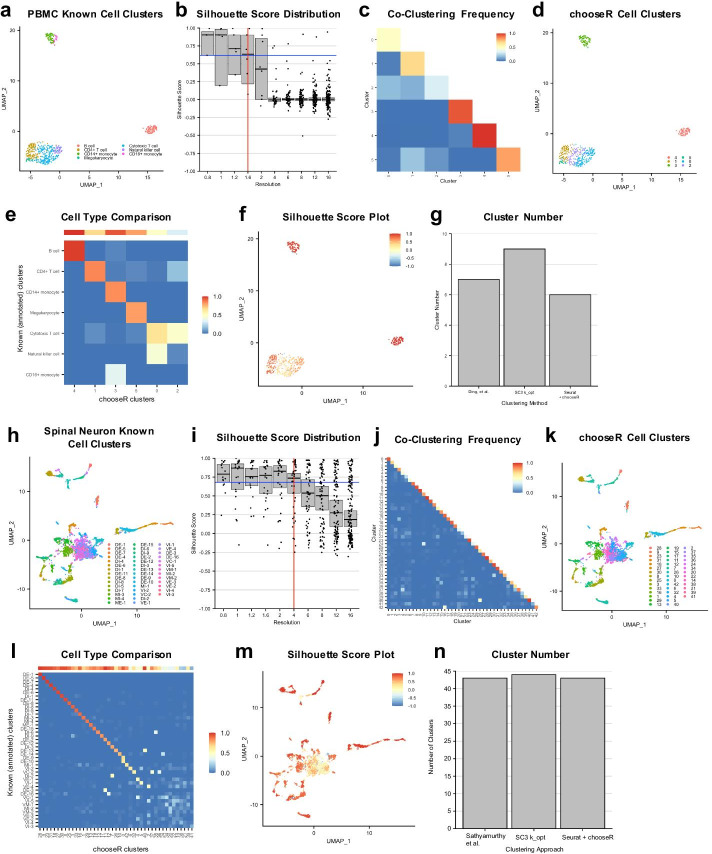
Fig. 3.
chooseR recovers annotated cell types in datasets of varying size and complexity. a–g Results of using chooseR and Seurat on the Ding et al. [18] Smart-Seq human PBMC dataset. a UMAP with cells colored by published, annotated clusters. b Silhouette distribution plot over multiple resolution parameter values, with each dot representing a cluster. Median with 95% CI is shown for each resolution. The vertical red line marks the optimal resolution, and the horizontal blue line marks the decision threshold (“Methods” section). The optimal parameter value (resolution) = 1.6. c Heatmap of average co-clustering frequency per cluster, following clustering on 100 random subsets of the data, each comprising 80% of the total cells. d UMAP displaying the optimal cluster labels at the chosen resolution. e UMAP displaying the per-cell silhouette scores at the chosen resolution. f Heatmap comparing the correlation between clusters at the optimal parameter value and known cell types, colored by the Dice coefficient. The horizontal bar at the top represents the within-cluster co-clustering frequency for each chooseR-derived cluster. g Bar plot comparing the number of recommended clusters across methods. h–n As in a–g but applied to the Sathyamurthy et al. [7] mouse spinal cord dataset