Abstract
Collagenous stromal accumulations predict a worse clinical outcome in a variety of malignancies. Better tools are needed to elucidate the way in which collagen influences cancer cells. Here, we report a method to generate collagenous matrices that are deficient in key post-translational modifications and evaluate cancer cell behaviors on those matrices. We utilized genetic and biochemical approaches to inhibit lysine hydroxylation and glucosylation on collagen produced by MC-3T3-E1 murine osteoblasts (MC cells). Seeded onto MC cell-derived matrix surface, multicellular aggregates containing lung adenocarcinoma cells alone or in combination with cancer-associated fibroblasts dissociated with temporal and spatial patterns that were influenced by collagen modifications. These findings demonstrate the feasibility of generating defined collagen matrices that are suitable for cell culture studies.
Abbreviations: Hylald, Aldehide Hydroxylysine; Lysald, Aldehide Lysine; DHLNL, Dehydro-dihydroxylysinonorleucine/its ketoamine; HLNL, Dehydro-hydroxylysinonorleucine/its ketoamine; ER, Endoplasmic Reticulum; G, Galactosyl group; GG, Glucosylgalactosyl group; PGGHG, Glucosylgalactosylhydroxylysine glucosidase; hLys, Helical domain Lysine; Hyl, Hydroxylysine; HLCCs, Hydroxylysine aldehyde-derived collagen cross-links; Hyp, Hydroxyproline; Lys, Lysine; LCC, Lysine aldehyde–derived cross-links; LH, Lysyl hydroxylases; LOX, Lysyl oxidases; PTMs, Post-translational modifications; Pro, Proline; tLys, Telopeptidyl Lysine
Keywords: Collagen, Collagen cross-links, Co-culture models, Lysyl hydroxylases, Lung cancer, Metastasis
Highlights
-
•
Feasibility of culturing multicellular aggregates on matrices with defined collagen modifications.
-
•
Collagen modifications influence cancer cell behavior.
-
•
This methodology is a useful tool for cancer researchers.
Introduction
Collagen is the most abundant protein in humans and the predominant structural protein in the extracellular matrix. Fibrillar collagens such as types I, II, III, V and XI are synthesized as precursors, procollagens, consisting of a 300 nm-long central triple-helical domain and, at amino (N)- and carboxy (C)-terminal ends, short, non-helical “telopeptides” and globular pro-peptides [1,2]. Type I collagen, the most abundant collagen, is composed of three polypeptide chains, two α1 and one α2, forming a right-handed triple helical structure. After secretion and proteolytic release of pro-peptide extensions by extracellular matrix metalloproteases, type I collagen molecules are packed into fibrils and stabilized by covalent intermolecular cross-links that allow collagen fibrils to perform mechanical functions [1]. During the biosynthesis, α1 and α2 chains acquire a number of functionally important post-translational modifications (PTMs). For instance, in the endoplasmic reticulum (ER), proline (Pro) residues can be hydroxylated to produce two forms of hydroxyproline (Hyp), i.e. 4-Hyp and 3-Hyp, in the sequences of X-(Pro)-Glycine (Gly) and (Pro)-4Hyp-Gly, respectively. The former is the predominant modification form and is critical for the stability of the collagen triple helix [2,3]. The latter, though very minor, may play a role in fibril assembly [4,5]. Another important collagen PTM include a series of lysine (Lys) modifications that occur in the intra- and extracellular compartments. In the ER, Lys residues within the helical domain sequence X-Lys-Gly (hLys) can be converted to hydroxylysine (Hyl) by lysyl hydroxylase 1 (LH1). The hydroxyl group of specific Hyl residues can then serve as a site for O-linked tandem galactosylation and glucosylation. Hyl is galactosylated by GLT25D family members (GLT25D1/2) [6], and galactosyl-Hyl is glucosylated by LH3 [7]. Telopeptidyl Lys (tLys) residues can be hydroxylated only by LH2 to generate tHyl [8,9]. The extent of Lys hydroxylation both in the helical and telopeptide domains significantly varies depending on the collagen types, tissues, and physiological conditions [1]. In the extracellular space, both tLys and tHyl residues in telopeptides can be oxidatively deaminated by lysyl oxidases (e.g. LOX and LOX-like/LOXL family members) to form aldehydes (Lysald and Hylald) that initiate collagen cross-linking [1,8].
Lys PTMs are essential for the structural functions of extracellular collagen matrix. For example, Hylald residues lead to the formation of stable cross-links called hydroxylysine aldehyde-derived collagen cross-links (HLCC) that produce the stiff matrix found in skeletal tissues such as bone and cartilage, whereas non-hydroxylated Lysald residues lead to less stable cross-links called lysine aldehyde–derived cross-links (LCC) found in soft connective tissues including skin and cornea [1]. Glycosylation of hLys residues regulates collagen fibrogenesis and cross-link maturation and stability [1]. Inactivating mutations in LH isoforms lead to connective tissue diseases, whereas aberrantly high expression of LHs contributes to osteoporosis, cancer, and fibrotic diseases [1,10,11].
Models have been developed to assess how collagen influences cellular behaviors. Three-dimensional gels containing extracted type I collagen, synthetic collagen matrices, and dermal explants have been utilized [[12], [13], [14], [15], [16]], but the collagen PTMs in these models vary depending on the extraction procedures used and the tissues from which they are derived. MC3T3-E1 murine calvaria-derived osteoblasts (MC cells) have been used to elucidate the enzymatic basis for collagen PTMs and how those PTMs influence collagen cross-linking, fibrillogenesis, and mineralization [7,17,18]. MC cells have high expression of type I collagen and all LH family members [9,18,19], but Lys residues on collagen are only partially hydroxylated and can be modulated by LH gain- and loss-of-function studies. Collagen cross-links generated by MC cells are formed and mature with predictable kinetics [20,21]. The cross-linked collagenous matrix produced by MC cells can be modified by subjecting them to genetic manipulation of collagen modifying enzymes [7,21,22]. For example, short hairpin RNA (shRNA)-mediated depletion of LH2 results in collagen matrices with reduced concentrations of HLCCs [22], whereas LH3 depletion results in matrices with less glucosylation [7,21]. When treated with the LOX inhibitor BAPN, MC cells produce a cross-link-deficient collagen matrix that influences osteoblastic differentiation [23]. Here, we show that MC cell-derived matrices can be used as tools to identify collagen post-translational modifications that influence cancer cells.
Results
Matrix preparation
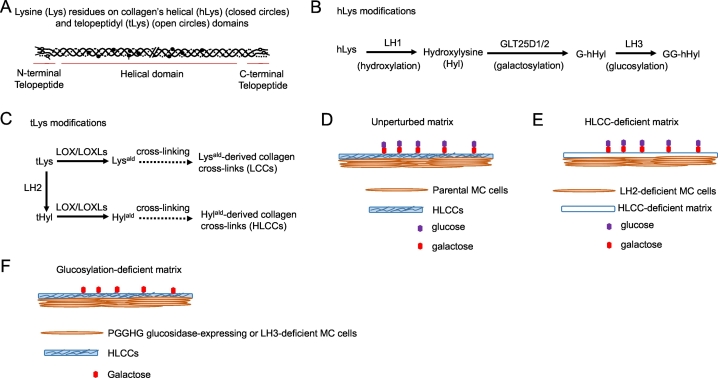
Based on fibrillar collagen's domain structure (Fig. 1A) and the post-translational modifications that occur on its hLys (Fig. 1B) and tLys (Fig. 1C) residues, we utilized short hairpin RNAs (shRNAs) and/or CRISPR/Cas-9-based approaches to generate LH2- and LH3-deficient MC cells that produce HLCC- and glucosylation-deficient collagen matrices, respectively (Fig. 1D-F, Supplementary Fig. 1A and B). Additionally, we generated glucosylation-deficient collagen matrices by ectopic expression of the collagen glycosidase protein-glucosylgalactosylhydroxylysine glucosidase (PGGHG) [24] in MC cells and by treating MC cell-derived matrices with PGGHG recombinant proteins (Fig. 1F, Supplementary Fig. 1C and D).
Fig. 1.
Collagen's structure and post-translational modifications and experimental design to inhibit those modifications. (A) Collagen's domain structure and lysine residues (arrows) on the helical domain (filled circles) and telopeptidyl domain (empty circles). (B, C) Enzymatic modifications of helical lysines (hLys) (B) and telopeptidyl lysines (tLys) (C). (D-F) MC cell-derived collagen matrix model. Unperturbed matrix (D), HLCC-deficient matrix (E), and glucosylation-deficient matrices (F) produced by parental and genetically modified MC cells.
Collagenous matrices were prepared for biochemical analysis and co-culture studies by seeding MC cells on 150-mm plastic plates and 24-well glass-bottom plates, respectively. Once MC cells reached 100% confluence, ascorbic acid (100 μg/ml) was added to stimulate collagen production. Typically, collagen matrix accumulated by seven days and extended throughout the 3-dimensional matrix by 14–21 days. With more prolonged incubation, matrices sometimes detached and became unusable.
Matrix visualization and characterization
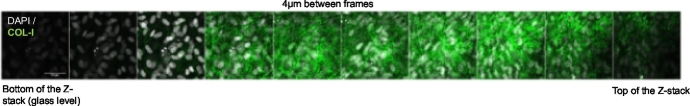
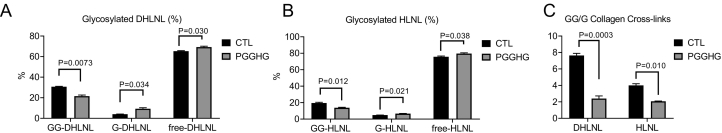
After fixation and staining with antibodies against type I collagen, matrices can be visualized as single-plane images or z-stacks (Fig. 2, Supplementary Fig. 2, and Supplementary Movie 1). Qualitative assessment of collagen abundance by collagen I staining and collagen structure by second harmonic generation microscopy demonstrated no appreciable changes induced by depletion of LH2 or LH3 (Supplementary Figs. 2 and 3). Using acid- and base-hydrolysis, glycosylation of two major reducible cross-links produced by MC cells at day 14, i.e. dehydro-dihydroxylysinonorleucine/its ketoamine (DHLNL) and dehydro-hydroxylysinonorleucine/its ketoamine (HLNL), were characterized by quantifying total, glucosylgalactosyl- (GG), galactosyl- (G), and free/non-glycosylated forms dihydroxylysinonorleucine (DHLNL) and hydroxylysinonorleucine (HLNL) in MC cell-derived matrices as reported [7,21]. Relative to controls, ectopic PGGHG-expressing MC cells had reduced levels of GG-DHLNL and GG-HLNL, increased levels of G-DHLNL and G-HLNL, and reduced GG/G ratios (Fig. 3), which recapitulates patterns reported in LH3-deficient MC cells [7].
Fig. 2.
Collagen deposition within 3-dimensional MC cell-derived matrix. Montage of a confocal Z-stack from bottom (left) to top (right) of the stack (4 μm increments). Matrices were fixed and stained with anti-type I collagen antibody, and DAPI for MC cell nuclei visualization. Collagen (green). DAPI (white). Scale bar size, 50 μm.
Fig. 3.
Quantification of collagen cross-links in MC cell-derived collagen matrices. Glucosylgalactosyl- (GG), galactosyl (G)-, and free dihydroxylysinonorleucine (DHLNL) and hydroxylysinonorleucine (HLNL) were quantified in MC cell-derived matrices produced by empty vector (control)- and ectopic PGGHG-expressing MC cells. Results are expressed as percentage of total cross-link. n = 3 replicates/condition.
Generation and seeding of multicellular aggregates
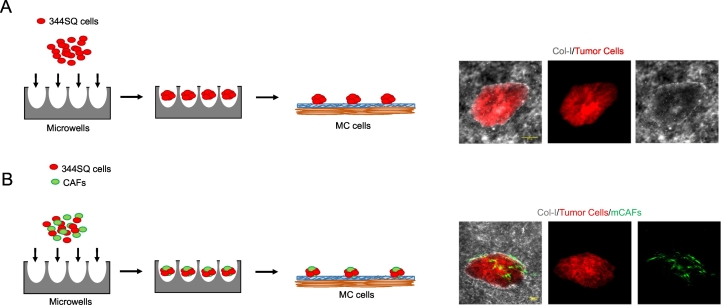
Multicellular aggregates of uniform cell number and consisting of one or more cell types in defined ratios can be generated in laser-ablated microwells (Fig. 4) [25]. The aggregates were collected two days after seeding into the microwells, layered onto the surface of MC cell-derived collagen matrices, and incubated for desired time periods.
Fig. 4.
Aggregate generation and seeding on MC cell-derived collagen matrices. (A-B) Schematic of multicellular aggregates containing RFP-tagged 344SQ cells alone (A) or in combination with GFP-tagged CAFs (B) generated in laser-ablated microwells (left). Fluorescent micrographs acquired with a standard fluorescent microscope (top) or a confocal microscope (bottom) of multicellular aggregates on parental MC cell-derived matrices stained with anti-type I collagen antibody 3 d after seeding (right). Collagen (white). 344SQ cells (red). CAFs (green). Scale bar size, 100 μm.
Multicellular aggregate time-to-flattening
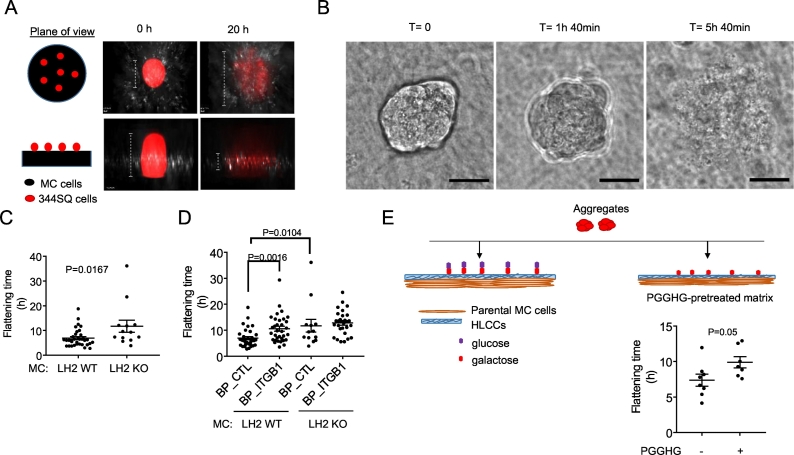
Utilizing a wide-field microscope equipped with a stage-top incubator and a 20× DIC objective that allows imaging of a wide field, multicellular aggregates were imaged beginning two to four hours after seeding, for a total duration of 24–48 h, taking images at 15 min intervals. Multicellular aggregates dissociated and flattened over time (Fig. 5A, B and Supplementary Movie 2). Time-to-flattening, which is defined as the time it takes for an aggregate to dissociate into a single-cell layer, is quantified by imaging aggregates at a z-plane that detects aggregate depth. Frames were manually analyzed to determine the time at which the aggregate boundary has completely blended with the underlying collagen matrix. Relative to controls, time-to-flattening was prolonged on HLCC-deficient matrices (Fig. 5C) and on HLCC-replete matrices treated with integrin-β1 blocking peptides (Fig. 5D), indicating that flattening is influenced by the type of collagen cross-link and anchoring to collagen receptors [26,27]. Similarly, time-to-flattening was prolonged on MC cell-derived matrices that had been pretreated with recombinant PGGHG protein (Fig. 5E).
Fig. 5.
Multicellular aggregate time-to-flattening assay. (A) Schematic of aggregate flattening over time. Cartoon representation of the plane of view (left). Fluorescent images of multicellular aggregates containing RFP-tagged 344SQ cells at the time of seeding (T = 0, center) and after seeding (T = 20 h) (right) on control shRNA-transfected MC cell-derived collagen matrices, stained with anti-type I collagen antibody (white). RFP-tagged 344SQ cells are visualized in the red channel. MC cells are not visualized. (B) DIC images depicting aggregate flattening over time. Loss of aggregate borders was used as a proxy for flattening. Scale bar size, 30 μm. (C) Dot plot of time-to-flattening of each multicellular aggregate (dot) on matrices generated by LH2 KO or wild-type (WT) MC cells. Two wells/condition, two movies/well. n ≥ 13 aggregates/condition (D) Dot plot of time-to-flattening of each multicellular aggregate (dot) on matrices generated by LH2 KO or wild-type (WT) MC cells in the presence of integrin-β1 blocking peptide (BP_ITGB1) or control peptide (BP_CTL). Two wells/condition, two movies/well. n ≥ 13 aggregates/condition (E) Cartoon of multicellular aggregates containing 344SQ cells seeded onto MC cell-derived treated matrices (top). Dot plot of time-to-flattening of each multicellular aggregate (dot) on MC cell-derived collagen matrices that had been pretreated for 4 h with either wild-type (+) or D300E-mutant (−) PGGHG recombinant protein. Two wells/condition, two movies/well. n ≥ 7 aggregates/condition.
Multicellular aggregate circularity
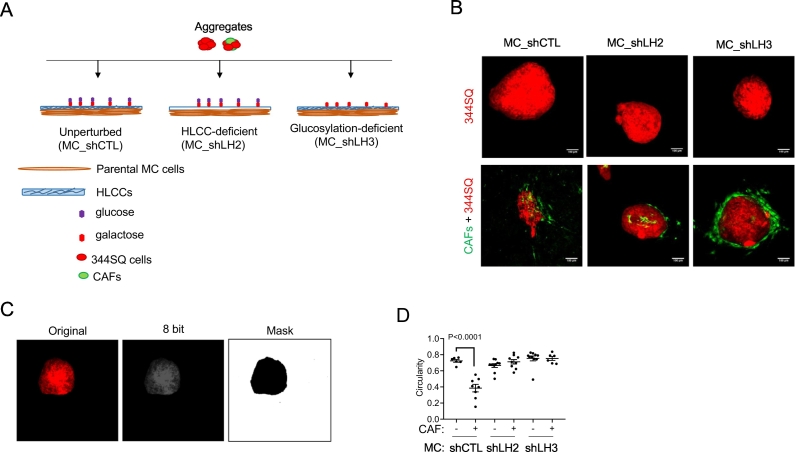
Seeded on the surface of collagen matrices, multicellular aggregates containing cancer cells and cancer-associated fibroblasts (CAFs) develop polarized features [28,29]. To detect CAF-induced aggregate polarization, multicellular aggregates containing 344SQ cells alone or in combination with CAFs (50:30 ratio) were seeded onto the surface of collagen matrices produced by LH2- or LH3-deficient or –replete MC cells (Fig. 6A), fixed, and visualized under a confocal microscope. Cancer cells and CAFs labelled with distinct fluorophores (Fig. 6B) were utilized to accurately define aggregate constituents and boundaries. Single-plane images generated from maximum projections of Z-stacks were transformed to eight-bit and subjected to thresholding to generate a mask of tumor cells within the aggregate (Fig. 6C), which then underwent circularity analysis. By utilizing loss of circularity as a proxy for polarization, we found that loss of LH2- or LH3-dependent collagen modifications impaired CAF-induced aggregate polarization (Fig. 6D).
Fig. 6.
Multicellular aggregate circularity assay. (A) Schematic of multicellular aggregates containing 344SQ cells alone or in combination with CAFs seeded on defined MC cell-derived collagen matrices. (B) Fluorescent confocal micrographs of multicellular aggregates taken three days after seeding on matrices generated by MC cells transfected with control shRNA (shCTL), LH2 shRNA (shLH2), or LH3 shRNA (shLH3). Scale bar size, 100 μm. (C) Representative images of image processing for circularity quantification. Fluorescent image of 344SQ cells in multicellular aggregate (left) is converted into eight-bit image (center) and thresholded to generate a mask (right) suitable for shape-descriptor analysis. (D) Dot plot of circularity values of each multicellular aggregate (dot) containing 344SQ cells alone or in combination with CAFs seeded on matrices generated by shCTL-, shLH2-, shLH3-transfected MC cells. Two wells/condition, n = 10 aggregates/condition.
Discussion
Intra-tumoral collagen accumulation is correlated with advanced disease and a shorter survival in multiple tumor types [[30], [31], [32]]. In one working model, collagen fibers activate collagen receptors that stimulate cell motility and provide migratory routes for invasive cancer cells [32]. However, the way in which collagen Lys residue modifications influence cancer cell behavior remains unclear, and currently available models cannot address this question. Here we show that osteoblast-derived matrices can be used as tools to identify collagen modifications that influence cancer cells.
Although the focus of our paper is primarily technical in nature, MC cell-derived matrices contain a variety of proteins other than collagens that could influence cancer cell behaviors [[33], [34], [35]], and the findings presented here warrant additional studies to exclude collagen-independent mechanisms by which MC cells influence cancer cell behaviors, to explore the biochemical and biophysical bases for these observations, and to determine the translational relevance of these findings in cancer patients. Interpreting these findings requires consideration of several factors. Depletion of a single collagen modifying enzyme in MC cells may lead to unintended collagen modifications if that enzyme is part of a feedforward loop involving other collagen modifying enzymes and/or their chaperone complexes [36]. Furthermore, collagen, collagen modifying enzymes, and other factors produced by cancer cells could influence matrix modifications by MC cells [37]. Conversely, matrix-bound secreted factors produced by MC cells could influence cancer cells through collagen-independent mechanisms. Such confounding factors can be minimized but not entirely eliminated by experimental designs incorporating congenic pairs of MC cells that differ only on the basis of a single collagen modifying enzyme. Efforts undertaken herein to further reduce collagen-independent effects of MC cells by rendering them non-viable after collagen production led to loss of matrix integrity. Fully addressing these issues will require synthetic collagen matrices that faithfully recapitulate the structural and biochemical features of native collagen.
High expression of collagen modifying enzymes drives metastasis in preclinical models and is a predictor of shorter survival duration in multiple tumor types [38]. These findings warrant the development of selective antagonists of collagen modifying enzymes. Given the scalability of MC cells in culture and multicellular aggregates in microwell plates, the assay reported here is potentially amenable to high throughput screening of small molecules and could serve as a secondary assay following the identification of initial hits from cell-free enzymatic screens.
Materials and methods
Reagents
We purchased fetal bovine serum (FBS), Dulbecco's minimal essential medium (DMEM), RPMI Media 1640, MEMα media without ascorbic acid (#A10490), and DAPI from Life Technologies; paraformaldehyde (#15714-S) from Electron Microscopy Sciences; glass-bottom 35-mm (#P35G-1.5-14-C) and 24-well plates (#P24G-1.5-13-F) from Mattek; penicillin-streptomycin (#30-002-CI) from Corning; pyruvate (#11360-70) from Gibco; puromycin (#ant-pr) from InvivoGene; purified NA/LE Hamster IgM, λ1 isotype control (BD553957), and purified NA/LE Hamster anti-rat CD29 (BD 555002) from Biosciences; LH3 and control shRNAs from Sigma; anti-LH3 antibody (60058-1-Ig) from Proteintech; anti-β-actin antibody (13E5; 4970) from Cell Signaling Technologies; anti-type I collagen antibody (Rabbit, 203,002) from MD Bioproducts; secondary Alexa-488 (anti-rabbit A-11034) and Alexa-647 (anti-rabbit A-21245) secondary antibodies from Life Technologies; and Ascorbic acid (A4544),from Sigma; Jetprime (114-15) from Polyplus transfection; qScript cDNA SuperMix (95048-025) from QuantaBio; RNeasy Mini Kit (74106) and Ni-NTA superflow resin (30450) from Qiagen; E. coli strain Rosetta (DE3, 70954-3) from Millipore; IPTG (I2481C100) from GoldBio. pET-28b is a gift from Dr. Craig Vander Kooi at the University of Kentucky. Primers for qPCR were as follows, from Sigma: LH3 primers CGTGTTTGTGGAGCAGCCTACT (forward) and GGCATCTGCAATATGAGGCTCG (reverse), RLP32 primers GGAGAAGGTTCAAGGGCCAG (forward) and TGCTC CCATAACCGATGTTTG (reverse), PGGHG RT-PCR primers were as follows, from Sigma: CTTTGACGTGGAGCAGAACTTCC (forward) and CGTCACAGCCTCGTAAATCTCC (reverse).
Cell culture
Lung adenocarcinoma cells generated from KrasLA1/+;Tp53R172H/+ mice (344SQ-RFP) [39] were cultured in RPMI-1640 with 10% FBS. Cells were kept at 37 °C in an incubator with a humidified atmosphere containing 5% CO2. Cells were passaged when they reached 80% confluence. GFP-tagged CAFs were isolated from KrasLA1 mice as described [29] and cultured in DMEM with 10% FBS, 1% pyruvate, and 1% penicillin-streptomycin. CAFs were passaged upon reaching 75% confluence and used for a maximum of 5 passages after thawing. MC cells and derivatives [7,21,30] were cultured in ascorbic acid-deficient MEMα supplemented with 10% FBS in a humidified atmosphere with 5% CO2 at 37 °C and were passaged upon reading 75% confluence. 293T cells were cultured in DMEM with 10% FBS in a humidified atmosphere with 5% CO2 at 37 °C and were passaged upon reading 75% confluence. All multicellular aggregate experiments were performed using a mix DMEM and RPMI in a 1:1 ratio.
Generation of MC cell transfectants
To generate ectopic PGGHG-expressing MC cells, PGGHG-expressing and control lentiviruses were produced in 293T cells using the pMD2.G and psPAX2 plasmids [40]. MC cells were infected with lentivirus particles collected from 293T cell conditioned medium samples. After 48 h, puromycin (10 μg/ml) was added and selection was continued for 4 weeks to generate mass populations of stable transfectants. To generate LH3-deficient MC cells, parental MC cells were infected with lentiviruses carrying LH3 shRNA or control shRNA that had been packaged in 293T cells using the pMD2.G and psPAX2 plasmids. After 48 h, the cells were selected with puromycin (10 μg/ml) for 2 weeks to generate LH3-deficient MC cells. The derivation of Plod2 knockout MC cells by CRISPR/Cas-9-mediated gene editing will be reported elsewhere (M. Yamauchi in preparation). Confirmation of LH2 depletion by qPCR and western blot analysis of LH2 shRNA-transfected MC cells was reported [22]. Derivation and characterization of MC cells subjected to CRISPR/Cas-9-mediated Plod2 KO will be reported separately (M. Yamauchi).
Multicellular aggregates
Multicellular aggregates were generated in a 24-well plate containing 1700 laser-ablated microwells per well as described [25]. Prior to seeding, the microwells were passivated with 0.05% pluronic acid for 1 h. 344SQ cells alone (85,000 per well) or in combination with CAFs (51,000 per well) were seeded in each well and cultured for 48 h to generate uniform aggregates, each containing 50 344SQ-RFP cells alone or 50 344SQ cells and 30 CAFs. After 2 d, the aggregates are collected in 1 ml of media, and medium samples containing aggregates were seeded onto the surface of MC cell-derived collagen matrices (50 to 100 ul per well), making sure that the media does not extend beyond the glass surface of the well. After 2 h, additional media were added to bring total volume to 1 ml per well.
MC cell-derived matrix collection for biochemical analysis
For matrix biochemical analysis, 2 × 106 MC cells were seeded on a 150 mm plastic dish. Upon reaching confluence, cells were cultured for 2 more weeks and maintained with 3-times-per-week change of ascorbic acid-deficient MEMα supplemented with 10% FBS and 100 μg/ml ascorbic acid. The cell/matrix layer was then washed with cold PBS, scraped, and collected by centrifugation at 10,000 rpm for 30 min and lyophilized. An aliquot of the dried material was hydrolyzed with 6N HCl and subjected to amino acid analysis to measure the collagen content and to characterize amino acid composition. An aliquot was reduced with standardized NaB3H4, hydrolyzed with 6N HCl or 2N NaOH and subjected to analyses for amino acids, reducible and non-reducible cross-links, and glycosylation of Hyl and cross-links (for details, see [10,41,42]). All cross-links and their glycosylated forms, precursor aldehydes were quantified as moles/mol of collagen (n = 3 replicates/condition).
Co-culture assay
MC cells were seeded on a glass-bottom 35-mm dish or 24-well plate at 37 °C and incubated until reaching >100% confluency, at which point they were treated with 100 μg/ml ascorbic acid for 2 weeks, replacing media every other day, at which point a collagen layer could be observed by direct observation. At that time, the aggregates were seeded onto the matrix surface and subjected to live-cell imaging (n = 2 wells/condition, n = 2 movies/well, n ≥ 7 aggregates/condition) or allowed to dissociate and flatten for 3 days, at which point they were fixed and subjected to fluorescence or confocal microscopy (Nikon A1) (n = 2 wells/condition, n ≥ 10 aggregates/condition). For integrin-β1 blocking peptide experiments, blocking and control peptides were used at a final concentration of 8 μg/ml. Peptides were added to the media with the aggregates at the time of seeding.
Matrix preparation for immunofluorescence analysis
To assess collagen production, MC cell-derived matrices were washed 3 times with 1× PBS containing calcium and magnesium to insure attachment of MC cells to the glass surface and subsequently fixed with 4% paraformaldehyde at 4 °C overnight or at room temperature for 2 h. Samples were washed 3 times with 1× PBS and stained with anti-collagen-I antibody (Rabbit, 1:1000) at 4 °C overnight and washed 3 times with 1× PBS. Secondary antibody (Alexa, anti-rabbit, 1:500) and DAPI were added at room temperature for 2 h. Samples were washed 3 times with 1× PBS, and 1 ml of 1× PBS was added as a final step before visualization.
Microscopy
Images of fixed multicellular aggregates on collagen matrices were acquired with a NikonA1 confocal microscope, 10× objective. After collecting z-slices, the volume was rendered into 2-dimensional projections by maximum intensity projection algorithm (ImageJ). Alternatively, images of fixed multicellular aggregates on collagen matrices were acquired with a wide-field fluorescence microscope (NikonA1), 20× objective, single-plane images. To detect collagen deposition in 3-dimensions, we followed Nyquist parameters.
To quantify circularity of fixed multicellular aggregates images, 344SQ cells and CAFs labelled with distinct fluorophores were utilized to accurately define aggregate constituents and boundaries. After acquiring Z-stacks, a single-plane image was generated with the “Max Projection” command and transformed to 8-bit and subjected to thresholding (Otsu) to generate a mask. Objects were selected with the “Magic Wand” command and circularity was measured with the “Measure” command utilizing “shape descriptors” option.
Live-cell imaging of aggregates on top of collagen matrices was performed utilizing an automated Nikon A1 wide-field fluorescence microscope (20× objective). Three fields per well were taken at intervals of 15 min for up to 24 h. Z-volumes were acquired with a mid-plane of focus set between aggregate mid-body and the collagen surface. Time-to-flattening was quantified manually by analyzing movies (frame by frame) to determine the time at which aggregate boundary has completely blended with the underlying collagen matrix.
Second harmonic generation (SHG) microscopy
SHG microscopy of MC cell-derived matrices was performed in a Leica SP8 Confocal/Multi-Photon Microscope (Leica Microsystems) using 20× (1.0 NA) water immersion lens. Samples were illuminated using a tunable, pulsed near-infrared Coherent Chameleon Ti:Sapphire laser line. Second harmonic signal from fibrillar collagen, which is half the wavelength of the illuminating light, was then collected into a non-descanned detector. Backscattered SHG of MC cell-derived matrices were collected in epi-illumination mode (with wide-open confocal pinholes) using appropriate dichroic at the objective back aperture. SHG signals were separated into one of the confocal PMT detectors and the amplified SHG signal was collected.
Quantitative real-time PCR (qPCR)
Trizol (Invitrogen) was utilized to isolate total RNA from the cells, according to the manufacturer's protocol. mRNA levels were analyzed using a SYBR Green–based system (Applied Biosystems) after reverse transcription with qScript cDNA SuperMix. LH3 mRNA levels were normalized based on mRNA for ribosomal protein L32 (Rpl32).
Western blotting
Cells were washed with 1× PBS buffer and lysed with cell lysis buffer, obtaining totalal proteins. Cell lysates were separated by SDS-PAGE gel, transferred onto a nitrocellulose transfer membrane. The membrane was incubated with primary antibody against LH3, and afterwards with a horseradish peroxidase–conjugated secondary antibody. Protein bands were visualized with Pierce ECL Western Blotting substrate.
Reverse transcriptase PCR (RT-PCR)
MC cells were seeded in a T25 flask. Upon 80% confluence, cells were lysed, and mRNA was extracted with RNeasy Mini Kit according to the manufacturer's instructions. 1.0 μg of total RNA was then reverse transcribed into cDNA using a qScript cDNA SuperMix kit. The resultant reverse transcription products were amplified with PGGHG primers by PCR. Results were normalized on the basis of L32 RNA levels.
Lentiviral particle isolation
HEK293T cells were seeded in a T25 flask. Upon reaching 70% confluence, HEK293T cells were transfected with lentivirus packaging vectors together with PGGHG-expressing or empty vector using jetPRIME according to the manufacturer's instructions. The medium was changed 16 h after transfection. Conditioned medium containing lentivirus particles was harvested 48 and 72 h after medium change.
Recombinant PGGHG proteins
Full-length murine PGGHG cDNA was cloned into a modified pET-28b (Novagen) vector using NheI and NotI cloning sites with standard PCR-based methods. This modified pET-28b vector has NheI inserted in the linker between His6 and Thrombin recognition site, which changes the amino acid linker from GS to AS. PGGHG was expressed in E. coli strain Rosetta (DE3). Cells expressing PGGHG were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16 h at 16 °C. Cells were collected, pelleted and then resuspended in binding buffer (20 mM Tris, pH 8.0, 200 mM NaCl and 15 mM imidazole). The cells were lysed by sonication and then centrifuged at 23,000g for 15 min. The recombinant PGGHG proteins (wild type or D300E inactive mutant) were purified with immobilized metal affinity chromatography, eluted with 300 mM imidazole pH 8.0, separated by SDS-PAGE and visualized using coomassie blue staining.
PGGHG proteins were diluted in PGGHG buffer (50 mM acetate buffer pH 5.3, 150 mM NaCl). MC cell-derived matrices were treated with 200ul of PGGHG buffer containing either PGGHG protein (0.02 mg/ml) or the D300E inactive PGGHG mutant (0.02 mg/ml) protein, for 4 h at 37 °C. Matrices were washed twice with RPMI:DMEM media. Multicellular aggregates were seeded onto the surface of PGGHG-modified collagen matrices and subjected to live-cell imaging for 24 h.
The following are the supplementary data related to this article.
Supplementary Fig. 1. Characterization of MC cells. (A) Quantitative RT-PCR (qPCR) analysis of LH3 RNA levels in MC cells stably transfected with control (shCTL) or LH3 (shLH3) shRNAs. (B) Western blot analysis of LH3 protein levels in shCTL- and shLH3-transfected MC cells. (C) RT-PCR analysis of PGGHG mRNA levels in MC cells stably transfected with PGGHG-expressing or empty (CTL) lentivirus. (D) Polyacrylamide gel electrophoresis of wild-type and inactive mutant recombinant PGGHG proteins.
Supplementary Fig. 2. Collagen I staining of MC cell-derived matrices. Confocal max projections of anti-collagen I antibody staining (green upper, white lower) and DAPI staining (white) to identify MC cell nuclei. Unmodified matrix (MC_shCTL), HLCC-deficient matrix (MC_shLH2) and glucosylation-deficient matrix (MC_shLH3). Scale bar: 100 μm. (n ≥ 3 wells/condition, n = 20 images/condition).
Supplementary Fig. 3. Second harmonic generation microscopy of MC cell-derived matrices. Confocal images showing collagen fiber organization in MC cell-derived matrices. Unmodified matrix (MC_shCTL), HLCC-deficient matrix (MC_shLH2) and glucosylation-deficient matrix (MC_shLH3). Scale bar: 100 μm. (n = 2 wells/condition).
MC cell-derived collagen matrix. Three-dimensional image of a confocal micrograph z-stack. Collagen I staining (green) and DAPI staining (white) to identify parental MC cell nuclei.
Aggregate flattening over time. DIC movie of a 344SQ aggregate seeded on top of matrix produced by parental MC cells. Loss of aggregate edges was used as a proxy for aggregate flattening. Scale bar size: 50 μm, 3 FPS.
CRediT authorship contribution statement
All the authors conceptualized the idea, performed experiments and participated in writing the manuscript.
Declaration of competing interest
J.M.K. has received consulting fees from Halozyme.
Acknowledgements
This work was supported by K99 1K99CA225633 (HFG), R01 CA181184 (JMK), R01 CA2111125 (JMK), and UT SPORE in Lung Cancer 600657-90-100300-53-MDGRN-00002293-0018 (JMK). JMK is The Gloria Lupton Tennison Distinguished Professor of Lung Cancer Research.
Contributor Information
Mitsuo Yamauchi, Email: Mitsuo_Yamauchi@unc.edu.
Jonathan M. Kurie, Email: jkurie@mdanderson.org.
References
- 1.Yamauchi M., Sricholpech M. Lysine post-translational modifications of collagen. Essays Biochem. 2012;52:113–133. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ricard-Blum S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011;3(1):a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., Weaver V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr Biol (Camb) 2015;7(10):1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyre D.R., Weis M., Hudson D.M., Wu J.J., Kim L. A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. J. Biol. Chem. 2011;286(10):7732–7736. doi: 10.1074/jbc.C110.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weis M.A., Hudson D.M., Kim L., Scott M., Wu J.J., Eyre D.R. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. J Biol Chem. 2010;285(4):2580–2590. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schegg B., Hülsmeier A.J., Rutschmann C., Maag C., Hennet T. Core glycosylation of collagen is initiated by two beta(1-O)galactosyltransferases. Mol. Cell. Biol. 2009;29(4):943–952. doi: 10.1128/MCB.02085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sricholpech M., Perdivara I., Nagaoka H., Yokoyama M., Tomer K.B., Yamauchi M. Lysyl hydroxylase 3 glucosylates galactosylhydroxylysine residues in type I collagen in osteoblast culture. J. Biol. Chem. 2011;286(11):8846–8856. doi: 10.1074/jbc.M110.178509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takaluoma K., Lantto J., Myllyharju J. Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix Biol. 2007;26(5):396–403. doi: 10.1016/j.matbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Uzawa K., Grzesik W.J., Nishiura T., Kuznetsov S.A., Robey P.G., Brenner D.A., Yamauchi M. Differential expression of human lysyl hydroxylase genes, lysine hydroxylation, and cross-linking of type I collagen during osteoblastic differentiation in vitro. J. Bone Miner. Res. 1999;14(8):1272–1280. doi: 10.1359/jbmr.1999.14.8.1272. [DOI] [PubMed] [Google Scholar]
- 10.Yamauchi M., Taga Y., Hattori S., Shiiba M., Terajima M. Analysis of collagen and elastin cross-links. Methods Cell Biol. 2018;143:115–132. doi: 10.1016/bs.mcb.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Pornprasertsuk S., Duarte W.R., Mochida Y., Yamauchi M. Overexpression of lysyl hydroxylase-2b leads to defective collagen fibrillogenesis and matrix mineralization. J. Bone Miner. Res. 2005;20(1):81–87. doi: 10.1359/JBMR.041026. [DOI] [PubMed] [Google Scholar]
- 12.Ashworth J.C., Thompson J.L., James J.R., Slater C.E., Pijuan-Galitó S., Lis-Slimak K., Holley R.J., Meade K.A., Thompson A., Arkill K.P., Tassieri M., Wright A.J., Farnie G., Merry C.L.R. Peptide gels of fully-defined composition and mechanics for probing cell-cell and cell-matrix interactions in vitro. Matrix Biol. 2020;85-86:15–33. doi: 10.1016/j.matbio.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haugh M.G., Murphy C.M., McKiernan R.C., Altenbuchner C., O’Brien F.J. Crosslinking and mechanical properties significantly influence cell attachment, proliferation, and migration within collagen glycosaminoglycan scaffolds. Tissue Eng Part A. 2011;17(9–10):1201–1208. doi: 10.1089/ten.TEA.2010.0590. [DOI] [PubMed] [Google Scholar]
- 14.Urbanczyk M., Layland S.L., Schenke-Layland K. The role of extracellular matrix in biomechanics and its impact on bioengineering of cells and 3D tissues. Matrix Biol. 2020;85-86:1–14. doi: 10.1016/j.matbio.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Wolf K., Alexander S., Schacht V., Coussens L.M., von Andrian U.H., van Rheenen J., Deryugina E., Friedl P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009;20(8):931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt H.C., Lewis D., Ciccaglione M., Connor S., Smith Q., Hickey J.W., Schneck J.P., Gerecht S. Collagen fiber structure guides 3D motility of cytotoxic T lymphocytes. Matrix Biol. 2020;85-86:147–159. doi: 10.1016/j.matbio.2019.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pornprasertsuk S., Duarte W.R., Mochida Y., Yamauchi M. Lysyl hydroxylase-2b directs collagen cross-linking pathways in MC3T3-E1 cells. J. Bone Miner. Res. 2004;19(8):1349–1355. doi: 10.1359/JBMR.040323. [DOI] [PubMed] [Google Scholar]
- 18.Terajima M., Taga Y., Sricholpech M., Kayashima Y., Sumida N., Maeda N., Hattori S., Yamauchi M. Role of glycosyltransferase 25 domain 1 in type I collagen glycosylation and molecular phenotypes. Biochemistry. 2019;58(50):5040–5051. doi: 10.1021/acs.biochem.8b00984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hata R., Hori H., Nagai Y., Tanaka S., Kondo M., Hiramatsu M., Utsumi N., Kumegawa M. Selective inhibition of type I collagen synthesis in osteoblastic cells by epidermal growth factor. Endocrinology. 1984;115(3):867–876. doi: 10.1210/endo-115-3-867. [DOI] [PubMed] [Google Scholar]
- 20.Kuboki Y., Kudo A., Mizuno M., Kawamura M. Time-dependent changes of collagen cross-links and their precursors in the culture of osteogenic cells. Calcif. Tissue Int. 1992;50(5):473–480. doi: 10.1007/BF00296780. [DOI] [PubMed] [Google Scholar]
- 21.Sricholpech M., Perdivara I., Yokoyama M., Nagaoka H., Terajima M., Tomer K.B., Yamauchi M. Lysyl hydroxylase 3-mediated glucosylation in type I collagen: molecular loci and biological significance. J. Biol. Chem. 2012;287(27):22998–23009. doi: 10.1074/jbc.M112.343954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Y., Guo H., Terajima M., Banerjee P., Liu X., Yu J., Momin A.A., Katayama H., Hanash S.M., Burns A.R., Fields G.B., Yamauchi M., Kurie J.M. Lysyl hydroxylase 2 is secreted by tumor cells and can modify collagen in the extracellular space. J. Biol. Chem. 2016;291(50):25799–25808. doi: 10.1074/jbc.M116.759803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ida T., Kaku M., Kitami M., Terajima M., Rosales Rocabado J.M., Akiba Y., Nagasawa M., Yamauchi M., Uoshima K. Extracellular matrix with defective collagen cross-linking affects the differentiation of bone cells. PLoS One. 2018;13(9) doi: 10.1371/journal.pone.0204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamazaki H., Hamazaki M.H. Catalytic site of human protein-glucosylgalactosylhydroxylysine glucosidase: three crucial carboxyl residues were determined by cloning and site-directed mutagenesis. Biochem. Biophys. Res. Commun. 2016;469(3):357–362. doi: 10.1016/j.bbrc.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Albritton J.L., Roybal J.D., Paulsen S.J., Calafat N., Flores-Zaher J.A., Farach-Carson M.C., Gibbons D.L., Miller J.S. Ultrahigh-throughput generation and characterization of cellular aggregates in laser-ablated microwells of poly(dimethylsiloxane) RSC Adv. 2016;6(11):8980–8991. doi: 10.1039/C5RA26022A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heino J. The collagen family members as cell adhesion proteins. Bioessays. 2007;29(10):1001–1010. doi: 10.1002/bies.20636. [DOI] [PubMed] [Google Scholar]
- 27.Leitinger B., Hohenester E. Mammalian collagen receptors. Matrix Biol. 2007;26(3):146–155. doi: 10.1016/j.matbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J.F., Harrington K., Sahai E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007;9(12):1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 29.D. Pankova, Y. Chen, M. Terajima, M.J. Schliekelman, B.N. Baird, M. Fahrenholtz, L. Sun, B.J. Gill, T.J. Vadakkan, M.P. Kim, Y.H. Ahn, J.D. Roybal, X. Liu, E.R. Parra Cuentas, J. Rodriguez, Wistuba, II, C.J. Creighton, D.L. Gibbons, J.M. Hicks, M.E. Dickinson, J.L. West, K.J. Grande-Allen, S.M. Hanash, M. Yamauchi, J.M. Kurie, Cancer-associated fibroblasts induce a collagen cross-link switch in tumor stroma, Mol Cancer Res 14(3) (2016) 287–95. [DOI] [PMC free article] [PubMed]
- 30.Y. Chen, M. Terajima, Y. Yang, L. Sun, Y.H. Ahn, D. Pankova, D.S. Puperi, T. Watanabe, M.P. Kim, S.H. Blackmon, J. Rodriguez, H. Liu, C. Behrens, Wistuba, II, R. Minelli, K.L. Scott, J. Sanchez-Adams, F. Guilak, D. Pati, N. Thilaganathan, A.R. Burns, C.J. Creighton, E.D. Martinez, T. Zal, K.J. Grande-Allen, M. Yamauchi, J.M. Kurie, Lysyl hydroxylase 2 induces a collagen cross-link switch in tumor stroma, J Clin Invest 125(3) (2015) 1147–62. [DOI] [PMC free article] [PubMed]
- 31.Eisinger-Mathason T.S., Zhang M., Qiu Q., Skuli N., Nakazawa M.S., Karakasheva T., Mucaj V., Shay J.E., Stangenberg L., Sadri N., Puré E., Yoon S.S., Kirsch D.G., Simon M.C. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3(10):1190–1205. doi: 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D.L., Weaver V.M. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139(5):891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garantziotis S., Savani R.C. Hyaluronan biology: a complex balancing act of structure, function, location and context. Matrix Biol. 2019;78-79:1–10. doi: 10.1016/j.matbio.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karamanos N.K., Theocharis A.D., Neill T., Iozzo R.V. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 2019;75-76:1–11. doi: 10.1016/j.matbio.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theocharis A.D., Karamanos N.K. Proteoglycans remodeling in cancer: underlying molecular mechanisms. Matrix Biol. 2019;75-76:220–259. doi: 10.1016/j.matbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Ishikawa Y., Boudko S., Bächinger H.P. Ziploc-ing the structure: triple helix formation is coordinated by rough endoplasmic reticulum resident PPIases. Biochim. Biophys. Acta. 2015;1850(10):1983–1993. doi: 10.1016/j.bbagen.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 37.Sprangers S., Everts V. Molecular pathways of cell-mediated degradation of fibrillar collagen. Matrix Biol. 2019;75-76:190–200. doi: 10.1016/j.matbio.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Yamauchi M., Barker T.H., Gibbons D.L., Kurie J.M. The fibrotic tumor stroma. J. Clin. Invest. 2018;128(1):16–25. doi: 10.1172/JCI93554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Padhye A., Ungewiss C., Fradette J.J., Rodriguez B.L., Albritton J.L., Miller J.S., Gibbons D.L. A novel ex vivo tumor system identifies Src-mediated invasion and metastasis in mesenchymal tumor cells in non-small cell lung cancer. Sci. Rep. 2019;9(1):4819. doi: 10.1038/s41598-019-41301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo H.F., Tsai C.L., Terajima M., Tan X., Banerjee P., Miller M.D., Liu X., Yu J., Byemerwa J., Alvarado S., Kaoud T.S., Dalby K.N., Bota-Rabassedas N., Chen Y., Yamauchi M., Tainer J.A., Phillips G.N., Jr., Kurie J.M. Pro-metastatic collagen lysyl hydroxylase dimer assemblies stabilized by Fe(2+)-binding. Nat. Commun. 2018;9(1):512. doi: 10.1038/s41467-018-02859-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamauchi M., Sricholpech M., Terajima M., Tomer K.B., Perdivara I. Glycosylation of type I collagen. Methods Mol. Biol. 2019;1934:127–144. doi: 10.1007/978-1-4939-9055-9_9. [DOI] [PubMed] [Google Scholar]
- 42.Yamauchi M., Terajima M., Shiiba M. Lysine hydroxylation and cross-linking of collagen. Methods Mol. Biol. 2019;1934:309–324. doi: 10.1007/978-1-4939-9055-9_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. 1. Characterization of MC cells. (A) Quantitative RT-PCR (qPCR) analysis of LH3 RNA levels in MC cells stably transfected with control (shCTL) or LH3 (shLH3) shRNAs. (B) Western blot analysis of LH3 protein levels in shCTL- and shLH3-transfected MC cells. (C) RT-PCR analysis of PGGHG mRNA levels in MC cells stably transfected with PGGHG-expressing or empty (CTL) lentivirus. (D) Polyacrylamide gel electrophoresis of wild-type and inactive mutant recombinant PGGHG proteins.
Supplementary Fig. 2. Collagen I staining of MC cell-derived matrices. Confocal max projections of anti-collagen I antibody staining (green upper, white lower) and DAPI staining (white) to identify MC cell nuclei. Unmodified matrix (MC_shCTL), HLCC-deficient matrix (MC_shLH2) and glucosylation-deficient matrix (MC_shLH3). Scale bar: 100 μm. (n ≥ 3 wells/condition, n = 20 images/condition).
Supplementary Fig. 3. Second harmonic generation microscopy of MC cell-derived matrices. Confocal images showing collagen fiber organization in MC cell-derived matrices. Unmodified matrix (MC_shCTL), HLCC-deficient matrix (MC_shLH2) and glucosylation-deficient matrix (MC_shLH3). Scale bar: 100 μm. (n = 2 wells/condition).
MC cell-derived collagen matrix. Three-dimensional image of a confocal micrograph z-stack. Collagen I staining (green) and DAPI staining (white) to identify parental MC cell nuclei.
Aggregate flattening over time. DIC movie of a 344SQ aggregate seeded on top of matrix produced by parental MC cells. Loss of aggregate edges was used as a proxy for aggregate flattening. Scale bar size: 50 μm, 3 FPS.