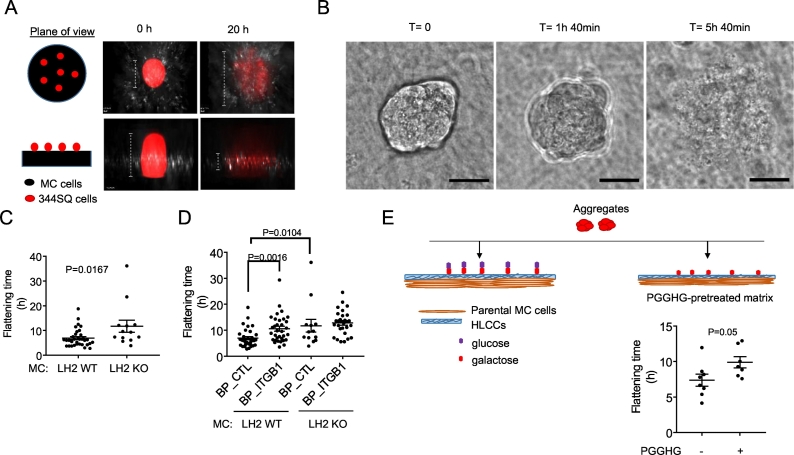
Fig. 5.
Multicellular aggregate time-to-flattening assay. (A) Schematic of aggregate flattening over time. Cartoon representation of the plane of view (left). Fluorescent images of multicellular aggregates containing RFP-tagged 344SQ cells at the time of seeding (T = 0, center) and after seeding (T = 20 h) (right) on control shRNA-transfected MC cell-derived collagen matrices, stained with anti-type I collagen antibody (white). RFP-tagged 344SQ cells are visualized in the red channel. MC cells are not visualized. (B) DIC images depicting aggregate flattening over time. Loss of aggregate borders was used as a proxy for flattening. Scale bar size, 30 μm. (C) Dot plot of time-to-flattening of each multicellular aggregate (dot) on matrices generated by LH2 KO or wild-type (WT) MC cells. Two wells/condition, two movies/well. n ≥ 13 aggregates/condition (D) Dot plot of time-to-flattening of each multicellular aggregate (dot) on matrices generated by LH2 KO or wild-type (WT) MC cells in the presence of integrin-β1 blocking peptide (BP_ITGB1) or control peptide (BP_CTL). Two wells/condition, two movies/well. n ≥ 13 aggregates/condition (E) Cartoon of multicellular aggregates containing 344SQ cells seeded onto MC cell-derived treated matrices (top). Dot plot of time-to-flattening of each multicellular aggregate (dot) on MC cell-derived collagen matrices that had been pretreated for 4 h with either wild-type (+) or D300E-mutant (−) PGGHG recombinant protein. Two wells/condition, two movies/well. n ≥ 7 aggregates/condition.