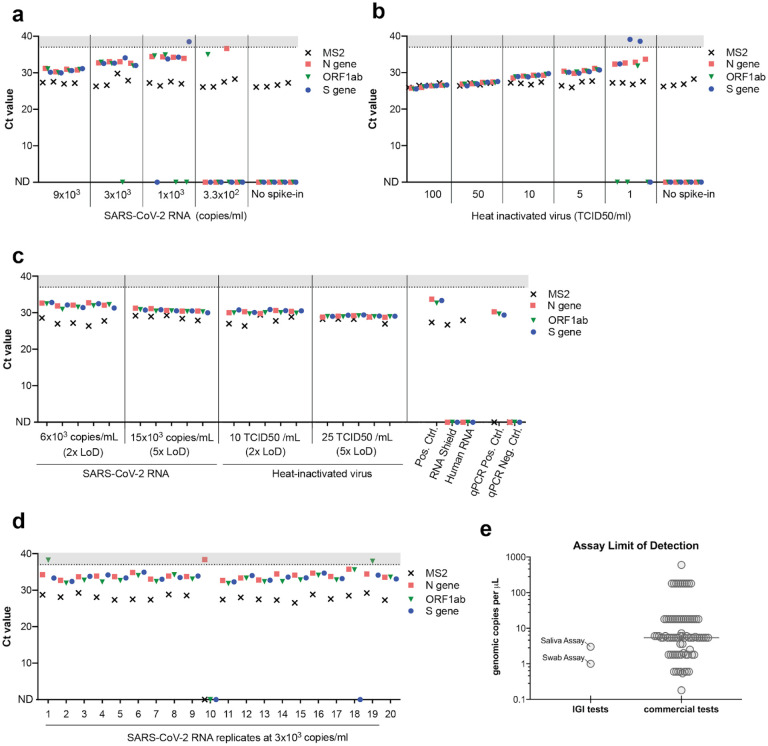
Figure 2: Validation of the IGI’s assay for detecting SARS-CoV-2 RNA from saliva —
Saliva collected from four unique donors (negative for SARS-CoV-2 by swab) was used to generate a titration curve of ThermoFisher COVID-19 Positive Control SARS-CoV-2 RNA (a) or heat-inactivated virus (b) to determine the assay’s limit of detection (LoD). c, SARS-CoV-2 RNA or heat-inactivated virus was spiked in at 2x or 5x the LoD into 20 unique saliva samples previously determined to be negative for SARS-CoV-2. d, SARS-CoV-2 RNA was spiked into 20 unique saliva samples previously determined to be negative for SARS-CoV-2 at 1x LoD (3×103 copies/ml). A positive extraction control (Pos. Ctrl) negative extraction controls (DNA/RNA Shield, Human RNA) and qPCR controls all returned expected results (c). Ct values >37 are shaded in gray. Undetected Ct values are plotted as zero and designated by “ND”, not detected. e, Limit of detection (RNA copies/μl) comparison of commercial assays and the IGI saliva and IGI swab tests generated from the FDA website10.