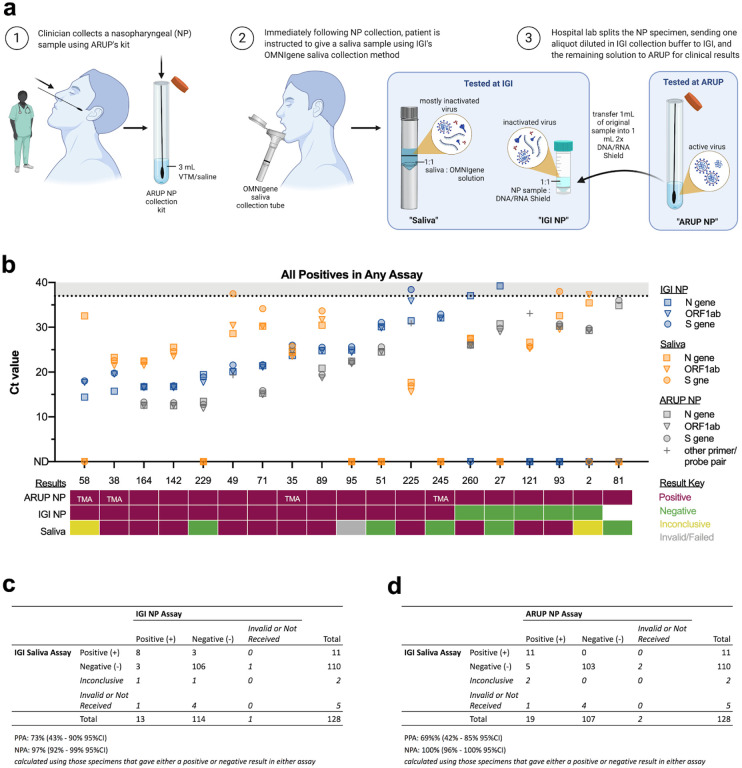
Figure 5: Clinical concordance between IGI-FAST saliva and NP swab samples —
The IGI partnered with Washington Hospital Healthcare System (WHHS) to collect paired nasopharyngeal (NP) swab and saliva specimens to assess the concordance between NP swab and saliva-based tests for detection of SARS-CoV-2. a, A schematic of how paired samples were collected and split for analysis. Note that undiluted NP samples were analyzed at ARUP Laboratories (ARUP NP) whereas NP samples analyzed at IGI (IGI NP) were diluted 1:1 with DNA/RNA Shield prior to extraction. b, Viral Ct values for IGI NP, saliva samples, and ARUP (where available) (top) and the final result of ARUP NP, IGI NP, and saliva samples (bottom). TMA indicates the sample was analyzed using transcription-mediated amplification, thus no Ct values were generated. Samples were sorted by the IGI NP N gene Ct value. As only positive samples are presented, MS2 Ct values were omitted for clarity. Ct values >37 are shaded in gray. Undetected Ct values are plotted as zero and designated by “ND”, not detected. An aliquot of the NP sample for patient 81 was never received by IGI. c, Concordance between the IGI saliva and IGI NP and d, IGI saliva and ARUP NP assays. Note: some individual samples had no result (invalid or no sample/result received) and were thus left out of Positive Percent Agreement (PPA) and Negative Percent Agreement (NPA) calculations.