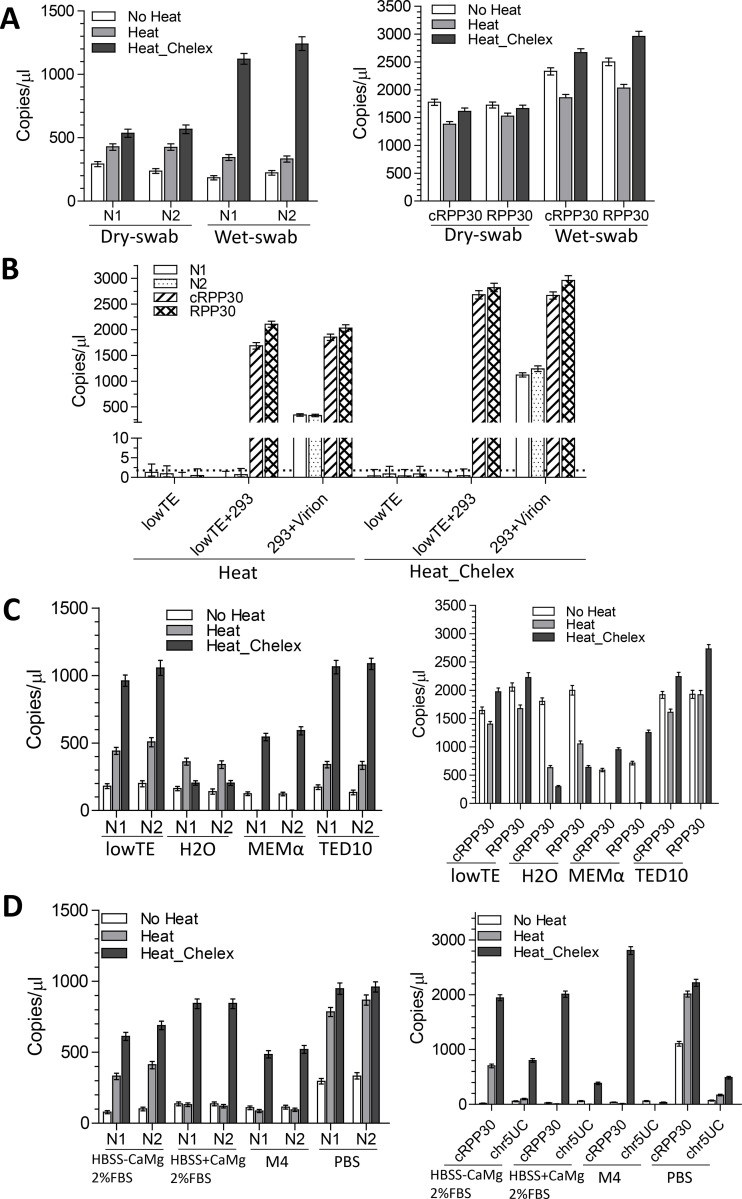
Figure 1.
RT-ddPCR assays for simulated dry- and wet-swab using RNA-extraction free methods for SARS-CoV-2 detection. (A) 200,000 SASRS-Cov-2 virions and 20,000 293FT cells were dried at room temperature in a speedvac and then resuspend in 200 μl lowTE buffer to mimic the dry swab. The same amount of virions and 293FT cells directly added to 200 μl lowTE buffer was used to mimic the wet swab. The samples expected to have 1000 virion genome copies/μl were then used directly for RT-ddPCR (No Heat), heated at 98 °C for 5 min (Heat), or heated with 5% Chelex (Heat_Chelex). The RT-ddPCR reactions were carried out in one well for N1 and cRPP30 and another well for N2 and RPP30. (B) Negative controls and virion samples prepared as wet swab. The mean genome copies/μl of N1 & N2 were less than 1.2 in negative controls without virions added. N1 & N2 target SARS-CoV-2. cRPP30 is specific for RPP30 cDNA, and RPP30 targets both genomic DNA and cDNA. Copies/μl refers to concentration in the samples used for RT-ddPCR. The error bars represent Poisson 95% confidence intervals. Dashed line indicates the threshold for the low detection limit of 1.8 copies/μl of SARS-CoV-2 virions. (C) Virions of 1000 genome copies/μl and 100 293FT cells/μl were prepared in lowTE, H2O, MEM alpha, or TED10, treated and assayed by RT-ddPCR as in (A). (D) Virions of 1000 genome copies/μl and 100 293FT cells/μl were prepared in HBSS with or without Ca2+ & Mg2+ supplemented with 2% FBS, M4, or PBS, treated and assayed by RT-ddPCR in for N1, N2, cRPP30 and chr5UC, a genomic DNA region on chromosome 5.