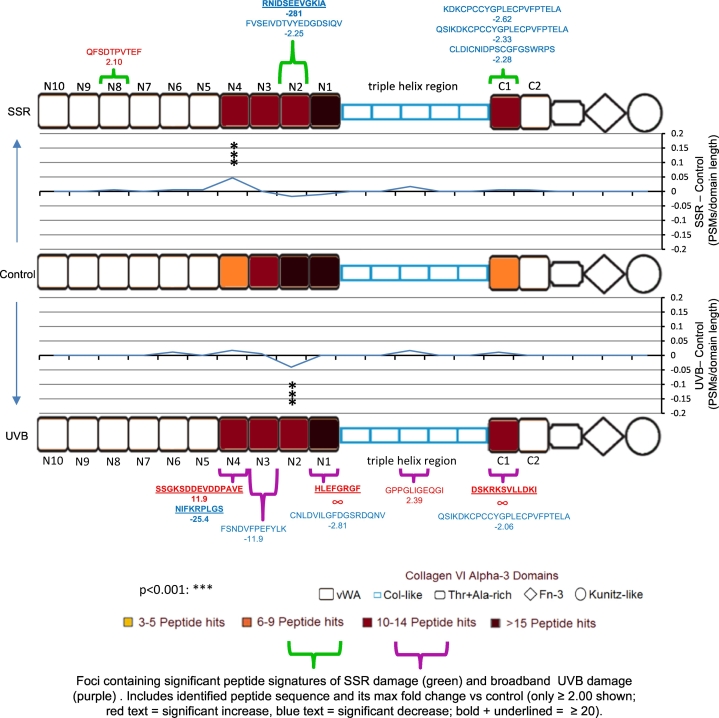
Fig. 6.
SSR and broadband UVB irradiation of HDF-derived microfibril isolations also leads to changes in the proteolytic susceptibility of specific protein regions within COL6A3. LC-MS/MS identified COL6A3 peptide sequences (PSMs: peptide prophet FDR ≤ 5%) were counted for each respective protein domain, normalised based on total spectrum count, averaged (N = 3) and subsequently heat mapped. Only domains containing an average of three peptides or more are shown. The PSM number corresponding to each broadband UVB- and SSR-irradiated COL6A3 domain were then subtracted from the counts of control and divided by the domain's primary sequence length to show regional fluctuations in proteolytic susceptibility (line graphs). Domains exhibiting significant differences in PSM numbers are also indicated (Bonferroni-corrected multiple comparisons tests taken from Fig. S3). Significantly different peptide sequences taken from Fig. 4 B and their fold changes are also mapped. Regional UVR damage to COL6A3 was limited compared to fibrillin-1, the highest (significant) changes in PSMs/domain length were seen within N4 (vWA) domain for SSR-irradiated and N2 domain for broadband UVB-irradiated. For both SSR and broadband UVB, data dependently quantified peptide sequences (which were significantly different in relative abundance) appear to cluster at vWA domains on the triple helix side of both globular regions of COL6A3: between N1 and N4 and at C1.