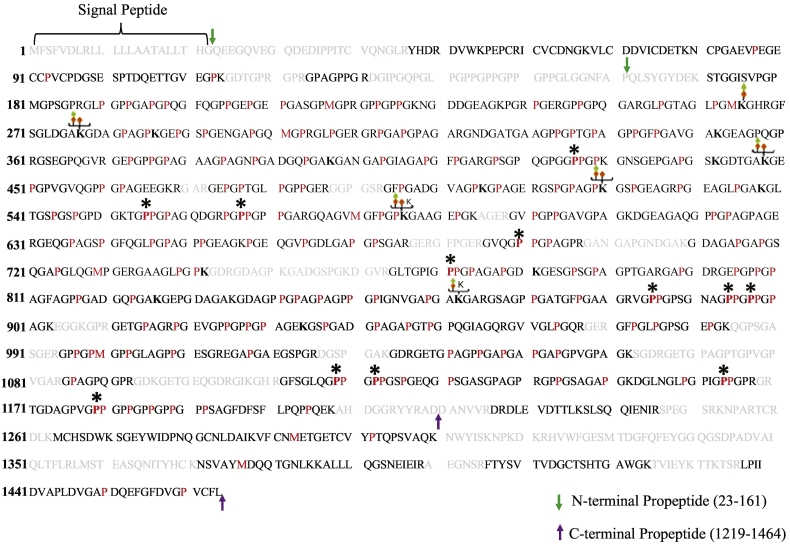
Fig. 7.
Comprehensive map of hydroxylation and O-linked glycosylation sites in human COL1A1. Lung fibroblast ECM was used to generate Collagen I PTM maps. MS identified collagen peptide sequences are shown in black which together represent an overall sequence coverage of 85% for the processed form of COL1A1. Signal peptide encompassing amino acid residues 1–22 are indicated. The green and purple arrows delimit the N-terminal (23–161) and C-terminal (1219–1464) pro-peptide sequences, respectively. Sequences not identified in this study are colored gray. “P” indicates 4-hydroxyproline occurring in the Yaa position of Gly-Xaa-Yaa motif, and a bold “P*” indicates 3-hydroxyproline in the Xaa position of Gly-Xaa-HyP motif. In addition, hydroxylation in red “P” residues occurring in the Xaa position followed by either Ala, Val, Met, Arg, Asp, or Glu in the Yaa position of Gly-Xaa-Yaa motif is reported but cannot be defined as either 3- or 4-HyP solely on the current MS/MS strategy [21]. Green and orange diamonds denote glucosyl and galactosyl sugar moieties attached to hydroxylysine (bold “K”). A summary of the PTMs is presented in Table 2, and PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figs. S3–S19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)