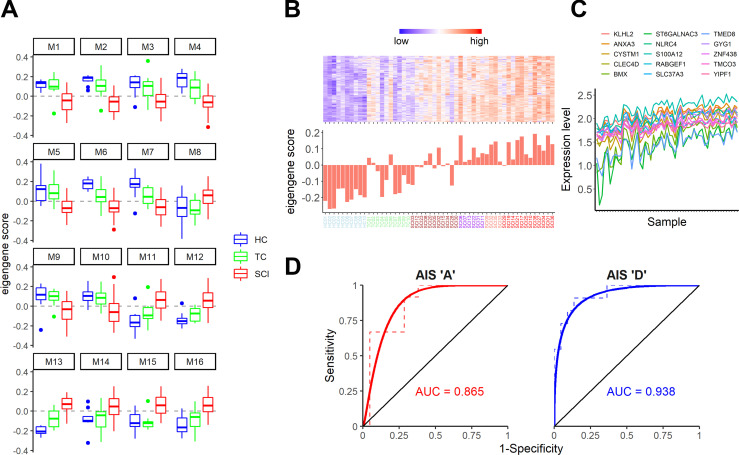
Figure 2.
Gene coexpression network analysis reveals transcriptional modules in peripheral WBCs that predict SCI severity. (A) Analysis of module eigengene (PC1) scores by patient cohort reveals 16 SCI-specific gene coexpression modules following unsupervised gene coexpression network analysis (one-way ANOVA, adjusted P value <0.05, Tukey’s P value < 0.05 for each comparison). Some modules (e.g., M4) display a gradual change in gene expression, whereas in others (e.g., M1, M5), HCs and TCs are very similar to each other but different from SCIs. n = 10 for HCs and TCs and 38 for SCIs. (B and C) The M13 module has the highest correlation to SCI severity (Spearman ρ = 0.82). In B is the heatmap of the top-seeded genes for this module (top), and the eigengene score for each one of the patients and controls (bottom). The graph in C shows the expression levels of the top 15 genes of the M13 module across all 58 samples. As expected from the analysis, these top genes of the module exhibit a strong coexpression pattern. (D) Receiver operating characteristic plots for the AIS A against the remaining SCIs (left) and the AIS D against the remaining SCIs (right). These plots show the strong predictive ability of our model for SCI patients with AIS A and D. The area under the curve (AUC) is 0.865 for A and 0.938 for D. n = 12 A vs. 21 SCIs and 11 D vs. 22 SCIs (color scheme in x-axis labels in B is as follows: blue = HC, green = TC, brown = AIS D, purple = AIS C, salmon = AIS B, and red = AIS A).