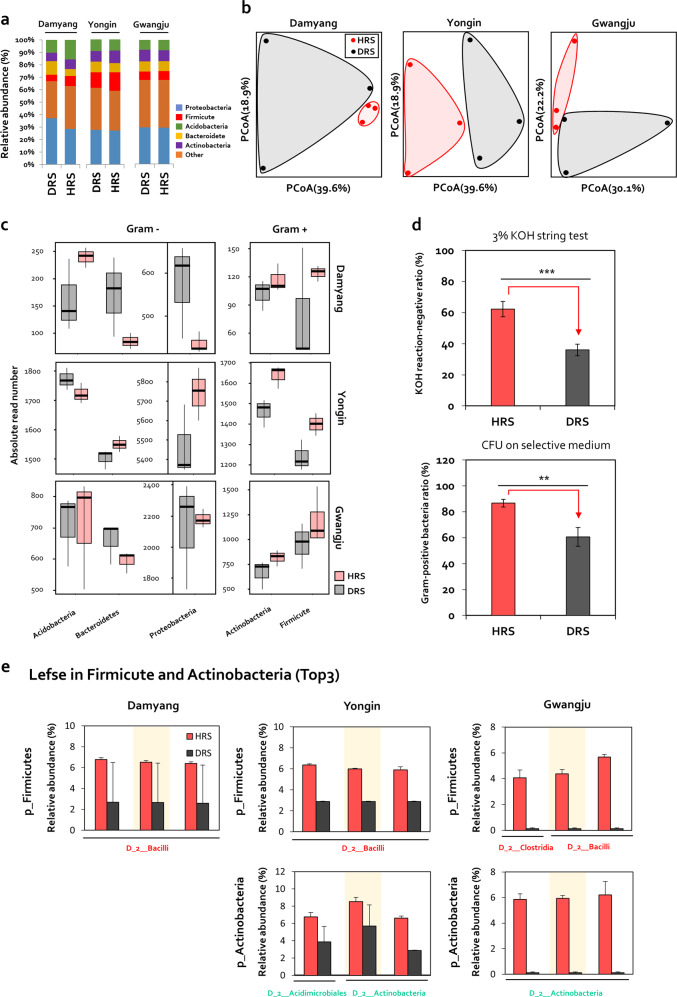
Fig. 2. Comparison of soil community structure between HRS and DRS samples based on pyrosequencing of 16S rRNA amplicons.
a Relative abundance of rhizobacteria at the phylum level in HRS and DRS samples collected from greenhouses in Damyang, Yongin, and Gwangju in South Korea. b Two-dimensional principal coordinate analysis (PCoA) of Bray–Curtis dissimilarity. Significant differences in microbial community composition were detected between HRS and DRS samples in Damyang, Yongin, and Gwangju. c The absolute read numbers of Firmicute, Actinobacteria, Proteobacteria, Acidobacteria, and Bacteroidetes in HRS and DRS from fields in Damyang, Yongin, and Gwangju. Gram+ Gram-positive bacterial groups, Gram− Gram-negative bacterial groups. d Measurement of the ratio of viable Firmicutes and Actinobacteria to Gram-negative bacteria in HRS and DRS samples using the 3% KOH string test and based on the quantification of colony-forming unit (CFU) values of bacterial isolates on TSA medium containing 20-μg/mL polymyxin B (toxic to Gram-negative bacteria) and 5-μg/mL vancomycin (toxic to Gram-positive bacteria). Data represent mean ± SEM. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001). e LefSe analysis of the Firmicutes and Actinobacteria community in HRS and DRS samples collected from Damyang, Yongin, and Gwangju. LefSe analysis was used to identify the most discriminating ASVs of Firmicutes and Actinobacteria phyla in HRS.