Abstract
Microcoleus vaginatus plays a prominent role as both primary producer and pioneer in biocrust communities from dryland soils. And yet, it cannot fix dinitrogen, essential in often nitrogen-limited drylands. But a diazotroph-rich “cyanosphere” has been described in M. vaginatus, hinting that there exists a C for N exchange between the photoautotroph and heterotrophic diazotrophs. We provide evidence for this by establishing such a symbiosis in culture and by showing that it is selective and dependent on nitrogen availability. In natural populations, provision of nitrogen resulted in loss of diazotrophs from the cyanosphere of M. vaginatus compared to controls, but provision of phosphorus did not. Co-culturing of pedigreed cyanosphere diazotroph isolates with axenic M. vaginatus resulted in copious growth in C and N-free medium, but co-culture with non-cyanosphere diazotrophs or other heterotrophs did not. Unexpectedly, bundle formation in M. vaginatus, diacritical to the genus but not seen in axenic culture, was restored in vitro by imposed nitrogen limitation or, even more strongly, by co-culture with diazotrophic partners, implicating this trait in the symbiosis. Our findings provide direct evidence for a symbiotic relationship between M. vaginatus and its cyanosphere and help explain how it can be a global pioneer in spite of its genetic shortcomings.
Subject terms: Microbial ecology, Soil microbiology
Introduction
Biological soil crusts (biocrusts) are complex communities of organisms that inhabit the surface of soils in arid and semiarid ecosystems (see [1, 2] for reviews), which account for nearly 45% of the terrestrial surface [3]. Biocrusts are well adapted to water limitation and intense solar radiation common to these ecosystems [4, 5], and depending on the local climate and soil textures, inhabitants of biocrust communities may include archaea [6], bacteria [7], algae [8], fungi [9], lichens [10], and bryophytes [11]. Biocrusts provide valuable ecosystem services that are closely linked to their successional stages [12–16]. Early-succession biocrusts are formed when bare dryland soils are colonized by non-heterocystous filamentous cyanobacteria, predominantly Microcoleus vaginatus (M. vaginatus), possibly the most abundant terrestrial cyanobacterium globally [17], but also other bundle-forming non-heterocystous filamentous cyanobacteria, like those in the less-known M. steenstrupii complex [18]. These cyanobacteria have the ability to secrete an exopolysaccharide sheath around groups of trichomes forming supracellular rope-like structures called bundles that can attach to soil particles and stabilize bare soils [19, 20]. Once stabilized, other organisms, such as heterocystous cyanobacteria, lichens, and bryophytes, can contribute to the maturation of the biocrust community, adding new ecosystem services [1].
The role of M. vaginatus as biocrust pioneer species in dryland ecosystems constitutes an ecological paradox, because it cannot fix nitrogen [21, 22] but dryland ecosystems are generally poor in nitrogen [23, 24] and barren soils within them even more so [25, 26], leaving an open question as to how this cyanobacterium accesses the nitrogen necessary for growth and colonization [27]. The presence of heterotrophic nitrogen-fixers in early successional biocrust had been predicted [28] and later confirmed [29], but only recently has their potential interaction with M. vaginatus been described [30]: In a recent study, it was demonstrated that M. vaginatus has a unique bacterial community associated to its bundles, a microscopic “cyanosphere” (by analogy to the rhizosphere). Not only does this cyanosphere community contain heterotrophic nitrogen-fixing bacteria, but its nitrogen-fixation potential is typically 100-fold higher than that of the surrounding biocrust soil [30]. Such spatial concentration of nitrogen-fixing heterotrophs suggests a potential resource trading relationship where fixed carbon is traded for fixed nitrogen. This finds support in the unusually high capacity of M. vaginatus to release photosynthate and its tendency to take up a large variety of compounds from the exometabolome [31, 32]. However, while bacterial communities in biocrusts have been characterized structurally [7], the precise functional roles of most heterotrophs have not.
An investigation of the proposed symbiotic resource trading relationship between M. vaginatus and the heterotrophic bacteria within its cyanosphere would thus be an important step in understanding the mechanisms of early successional biocrust formation and the relatively unstudied microbial ecology of these complex systems. It is reasonable to expect that nutrient availability may drive potential syntrophic interactions between M. vaginatus and its cyanosphere. If nutrient limitation is removed, one can also expect a release of the pressures to keep a diazotrophic cyanosphere, leading to a shift in cyanosphere community structure and function. Here, we test two hypotheses: (1) increased nitrogen availability will affect any existing resource trading relationship between M. vaginatus and its neighboring diazotrophic heterotrophs by disrupting the spatial organization of nitrogen-fixers in the cyanosphere, (2) heterotrophic nitrogen-fixing bacteria found within the cyanosphere of M. vaginatus are specifically suited to become resource trading partners. Two parallel approaches were developed to test these hypotheses. In one, we manipulated naturally occurring M. vaginatus-associated communities with nutrient additions while monitoring them structurally and functionally. In the other, we attempted to recreate the alleged symbiosis using co-cultures of the cyanobacterium with isolated cyanosphere heterotrophs under nitrogen limitation. Unexpectedly, our experiments also uncovered how conditions leading to the trading relationship induced M. vaginatus to form bundles, an ecologically important trait that may allow M. vaginatus to shape its cyanosphere.
Methods
Sampling location and procedures
Early successional biocrusts, specifically those composed mainly of Microcoleus spp., but not heterocystous cyanobacteria as determined by 16S rRNA community composition analysis, were sampled in February 2016 from the Fort Bliss military base, located in the Chihuahuan Desert (lat. 32.431069°, long. −105.984151°, El Paso, TX, USA) [33, 34]. Samples were taken using Petri dishes (15 cm diameter, 1 cm depth) to cut intact biocrust from the field [35]. Samples were air dried and maintained inactive at a low relative humidity (RH 15%) in darkness until experimentation in March 2017.
Cyanosphere community manipulation and Microcoleus bundle selection
In order to determine the effect of nitrogen availability on the hypothesized symbiosis between M. vaginatus and its diazotrophic cyanosphere, nutrient addition experiments were performed. Each 15 cm petri plate was divided evenly and distributed into three sterile 6-well plates (NUNC, Roskilde, Denmark). To relieve nitrogen limitation, 1 mL of a N solution (1.25 mM NH4Cl), diluted twofold from concentrations used for this purpose in cyanobacterial cultures [36], was added to six wells of one plate (n = 6). As a control, 1 mL of a P solution (1.25 mM K2HPO4) was added to six replicate wells in another plate. For another control, we added 1 mL of sterile reverse osmosis water (RO) to six wells in a third plate. We incubated all plates simulating the natural wetting/drying growth cycle in the field, with nine consecutive wetting and drying cycles, providing 48 h of hydration (and activity) followed by 24 h of desiccation (and stasis). Plates were incubated at 23 °C, under 18–20 μE m−2 s−1 of white light and with a 14 h illumination/10 h dark cycle. After incubation, individual bundles of M. vaginatus were pulled from the soil in the wells under the dissecting scope (see Supplementary Video in [30] for the exact procedure) and cleansed of excess soil by dragging bundles through sterile 2% agar plates [37]. Preliminary identification of the cyanobacteria in individual bundles was performed by microscopic observation. A total of 31 likely M. vaginatus bundles were selected for sequencing: nine from the N-addition treatment, ten from the P-addition, and twelve from the control (RO water).
Cyanosphere microbial community composition
Single-bundle DNA was extracted with a Powersoil extraction kit (QIAGEN, Hilden, Germany) using the standard protocol, with the exception of final DNA elution to 60 μL. Bacterial/Archaeal community analysis was performed via commercial next-generation sequencing in a MiSeq Illumina platform (Illumina, San Diego, CA, USA). Amplicon sequencing of the V4 region of the 16S rRNA gene was performed with barcoded primer set 515F/806R [38] following the Earth Microbiome Project protocol [39] for library preparation. PCR amplifications were done in triplicate, then pooled and quantified using a Quant-iT™ PicoGreen® dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Two hundred and forty nanograms of DNA of each replicate was pooled and cleaned using a QIA quick PCR purification kit (QIAGEN). The DNA in the pooled amplificate was quantified using an Illumina library Quantification Kit ABI Prism® (Kapa Biosystems, Wilmington, MA, USA) and diluted with NaOH to a final concentration of 4 nM, then denatured and diluted to a final concentration of 4 pM, and 30% of PhiX (Illumina) was added to the solution. The library was then loaded in the sequencer using the chemistry version 2 (2 × 250 paired end) and following manufacturer’s specifications [39]. Sequencing was performed in the Microbiome Analysis Laboratory at Arizona State University (Tempe, AZ, USA), yielding raw FASTQ sequence files.
Bioinformatic analysis
The raw FASTQ file was de-multiplexed within the MiSeq Illumina workflow under default parameters. Paired sequences were de-multiplexed and analyzed via Qiime2.10 [40], using the DADA2 plugin [41] where sequences were trimmed to include 250 bases from the V4 region, bound by 515F/806R primers [42], to create a feature table with representative sequences (features) and their frequency of occurrence. To remove highly variable positions, sequences were aligned with the MAFFT program [43]. FastTree [44] was used to generate a tree. Taxonomy was initially assigned with the Naive Bayes classifier trained on the Greengenes 13.8 release. Additional steps were taken to identify cyanobacteria, due to the poor and unreliable taxonomic resolution obtained with Greengenes [45]. Cyanobacterial sequences were filtered out from the feature table and those that attained >0.05% of the total number of cyanobacterial features were phylogenetically assigned using our own curated cyanobacteria database/tree version-0.22a (https://github.com/FGPLab/cydrasil/tree/0.22a) via RAxML [46] and displayed using ITOL [47]. Bundles with <80% of the cyanobacterial community composition attributable to M. vaginatus were discarded, leaving 17 bundles for downstream analysis; four control bundles, six for N-addition treatments and seven for P-addition treatments. For cyanosphere microbial community analyses, all M. vaginatus sequences were removed. Significance in compositional shifts was tested with permutational multivariate analysis of variance (PERMANOVA) calculated on Bray–Curtis similarity matrices of relative abundances derived from sequencing with 9999 permutations and visualized using 2-D NMDS plots. A similarity percentage analysis (SIMPER) was performed within the PRIMER software, v6 [48] to identify which community members contributed most to the dissimilarity (i.e., were differentially enriched) between treatments.
Assessment of nitrogen-fixation potential
In order to measure nitrogen-fixing potential of the cyanosphere communities, quantitative real-time PCR was used to quantify the gene copy number of 16S rRNA (a proxy for bacterial biomass) and nifH (a proxy for nitrogen-fixation ability) genes using the previously extracted genomic DNA, as previously described [30]. For 16S rRNA gene quantitation, a universal (bacterial/archaeal) primer set (338F 5′-ACTCCTACGG GAGGCAGCAG-3′, 518R 5′-GTATTACCG CGGCTGCTGG-3′) [49] was used. For nifH quantitation, a high coverage primer set for nifH (IDK3 5′-GCIWTHTAYG GIAARGGIGG IATHGGIAA-3′, DVV 5′-ATIGCRAAIC CICCRCAIAC IACRTC-3′) [50] was used. The PCR reactions were performed in triplicate using the Sso Fast mix (Bio-Rad, Hercules, CA, USA) in a ABI7900HT thermocycler (Applied Biosystems, Foster City, CA, USA) under conditions previously published [30, 51]. Nitrogen-fixation potential for each bundle was determined by calculating the ratio of nifH/16S gene copy numbers detected. Ratios were log transformed to comply with normality and variance homogeneity before testing for significance between nitrogen-fixation potential among treatments with a one-way ANOVA using R [52].
Targeted isolation of cyanosphere heterotrophs
Heterotrophic bacteria were isolated from biocrust using a variety of different enrichments and substrates, from which candidate bacteria were selected for co-culturing experiments. For the isolation of heterotrophic nitrogen-fixing bacteria, a modified version of a protocol for diazotroph isolation [53] was used. One gram of dried intact biocrust was rehydrated with 1 mL sterile RO water and incubated in culture room conditions (23 °C, 18–20 μE m−2 s−1, 14 h light cycle) for 24 h. Rehydrated biocrust was then added to 10 mL phosphate-buffered saline (PBS), vigorously shaken (120 rpm) for 30 m, and serial diluted in PBS. Dilutions of 10−3 were used for all diazotroph isolations. Nitrogen-free combined carbon (NFCC) [54, 55] medium was prepared, using 2 g/L of each of the following carbon sources: glucose, sucrose, trehalose, sodium pyruvate, and sodium acetate. NFCC plates (1% Gellan gum, 60 mm × 15 mm) were inoculated with 70 µL of biocrust serial dilution (10−3) and incubated in darkness under both aerobic (atmospheric O2) and microaerophilic conditions (2% O2 and 98% N2), at 23 °C for 20 days. Additional aerobic enrichments, following the same inoculation protocol in aerobic conditions using Burk’s medium [56] and BG110 + 1% sucrose [57] plates (1% gellan gum) were performed. In order to target diazotrophs residing in the cyanosphere specifically, M. vaginatus bundles were pulled from rehydrated biocrust and cleansed of excess soil as previously described [37]. Cleansed bundles were then placed on solid enrichment medium plates (Burk’s, BG110 + 1% sucrose) and incubated under standard conditions (23 °C, 18–20 μE m−2 s−1, 14 h light cycle) for 20 days, after which individual colonies and heterotrophic biomass near M. vaginatus bundles were picked based on morphological differences and streaked on new nitrogen-free media plates (NFCC, Burk’s, and BG110 + 1% sucrose where appropriate), repeating three times for each colony transfer, to obtain pure isolates.
Molecular identification of pure isolates and candidate selection
Genomic DNA was extracted from one colony of each heterotrophic isolate, resuspending biomass in 30 µL of sterile RO water in a sterile 0.5 mL PCR tube, microwaving for 30 s to lyse cells, and flicking tube to agitate. This process was repeated three times. The 16S rRNA gene was amplified using general bacterial primers 530F [58], and 1492R [59]. For a reaction volume of 21 µL, the following amounts were used: 9 µL Go Taq Mastermix 2× (Promega, Madison, WI, USA), 1 µL of each primer, 9 µL of H2O, and 1 µL of template DNA. The PCR conditions were as follows: 2 min of denaturation, followed by 35 rounds of temperature cycling (94 °C for 15 s, 55 °C for 30 s, 72 °C for 90 s) and a final extension at 72 °C for 7 min. Positive amplifications were Sanger sequenced at the ASU Genomics core facilities. Forward and reverse sequences were aligned in Geneious version 8.0 [60] and the 850–900 bp consensus sequences were submitted to basic local alignment search (BLAST) [61]. Taxonomy was initially assigned based off matches (>97% identity) to 16S rRNA sequences of strains in the NCBI database. Isolate sequences were then submitted to BLAST against the 16S rRNA sequence data from known cyanosphere bacteria as described in the literature [29, 30] and from sequencing data obtained in this study. Isolates matching either source were selected as candidates for co-culture experiments. Isolates were classified either as a typical members (or not) of M. vaginatus cyanosphere, and by their potential ecological function, as far as it was possible based on a literature search [62–71] (Table 1).
Table 1.
Bacterial isolates used in co-culture experiments with M. vaginatus PCC9802.
| Isolate information | Match to known cyanosphere members (% similarity to 16S rRNA gene) | ||||||
|---|---|---|---|---|---|---|---|
| Origin | Strain ID | Taxonomic assignment | Growth on N-free medium | nifH gene detected by PCR | Functional inference | Reference | |
| Cyanosphere associated | O80 | Arthrobacter sp. | + | + | Aerobic/saprophytic | [64] | 99% (this work) |
| METH4 | Massilia sp. | + | − | Aerobic/copiotrophic | [62, 65] | 98% (this work) | |
| O64 | Bacillus sp. | + | + | Strict or facultative aerobe/copiotroph | [63, 67] | 98% [29] | |
| Biocrust associated | S1 | Ensifer sp. | + | − | Aerobic, rhizobacteria | [68] | No match |
| S14 | Paenibacillus sp. | + | + | Copiotrophic, rhizobacteria | [63, 66] | No match | |
| B17 | Geodermatophilus sp. | + | − | Aerobic, rhizobacteria | [67] | No match | |
| METH2 | Streptomyces sp. | + | + | Aerobic/saprophytic | [63, 70] | No match | |
| Non-biocrust associated | K12 | Escherichia coli | − | − | Copiotrophic | [71] | No match |
| D1 | Serratia marcescens | − | − | Copiotrophic | [63, 71] | No match | |
Taxonomic assignment and functional inference based on BLAST of 16S rRNA gene sequence against the NCBI database. Cyanosphere match indicates isolate 16S rRNA sequence similarity to cyanosphere members detected with culture independent approaches here or elsewhere.
Nitrogen-fixation screening of heterotrophic isolates
To screen for nitrogen-fixation ability, isolates were subjected to PCR confirmation. The marker gene for nitrogen-fixation ability, nifH, was PCR amplified from isolate DNA using PolF/PolR [72] and IGK3/DVV [50] primer sets. For a reaction volume of 21 µL, the following amounts were used: 9 µL Go Taq Mastermix 2×, 1 µL of each primer, 9 µL of H2O, and 1 µL of template DNA. The PCR conditions were as follows: For PolF/PolR, 2 min of denaturation, followed by 30 rounds of temperature cycling (94 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min) and a final extension at 72 °C for 5 min. For IGK3/DVV, 10 min of denaturation, followed by 40 rounds of temperature cycling (95 °C for 45 s, 52 °C for 30 s, 72 °C for 40 s) and a final extension at 72 °C for 10 min. PCR products (6 µL) were observed on SYBR Safe (Invitrogen) stained 1% agarose gels.
Co-cultures
Heterotrophic isolates that matched cyanosphere 16S rRNA sequencing data were co-cultured with an axenic strain of M. vaginatus (PCC9802), originally isolated from biocrusts in the US southwest and available through the Pasteur Culture Collection of Cyanobacteria (Paris, France). M. vaginatus PCC9802 was maintained in 250 mL vented suspension culture flasks (Greiner Bio-One, Kremsmünster, Austria) in BG11 medium under standard conditions (23 °C, 18–20 μE m−2 s−1, 14 h light cycle). Non-diazotrophic heterotrophic strains atypical of biocrusts, as well as nitrogen-fixing isolates typical of biocrusts but not found in the cyanosphere were also used to serve as controls. All isolates used for co-culture experiments are listed in Table 1. To inoculate co-cultures, we harvested ten equivalently sized colonies of each heterotrophic isolate, resuspending them in 1 mL of nitrogen-free 10% BG110 medium. Both M. vaginatus and heterotrophs were washed three times prior to inoculation by pelleting cells in a centrifuge at 8000 rpm for 8 min, removing supernatant, and resuspending cells in 10% BG110 medium, to remove remnant N from the previous medium. The washed cyanobacteria and heterotrophic isolates were diluted ten-fold and combined before being plated on nitrogen-free medium (10% BG110) solidified with 1% Gellan gum (n = 3). Using Chl a as proxy for phototrophic biomass, final inoculation concentration for M. vaginatus was 0.7 ± 0.04 mg Chl a per plate. Chl a concentration was determined by adding 100 μL of homogenized cyanobacterial stock culture to 900 μL of acetone in 2 mL microcentrifuge tubes containing 0.25 g of 0.5 mm zirconium beads. Pigments were extracted by bead beating acetone/culture mixture for 2 min at 30 s intervals, and then storing in the dark for 24 h at 4 °C. Absorbance spectra and OD600 were recorded on a UV–visible spectrophotometer (Shizmadzu UV-1601, Kyoto, Japan). Co-cultures were incubated at 23 °C, 18–20 μE m−2 s−1, 14 h light cycle for 20 days before final inspection and analysis. After final inspection, co-culture plates were photographed and analyzed by importing images into Image J [73]. The software was then used to calculate percent cover of cyanobacterial biomass in the 56.8 cm2 area of each replicate plate (n = 3). Differences in average percent cover among co-cultures were tested for significance with a one-way ANOVA using R [52], after performing an arcsine square root transformation to maintain normality and equal variance required for parametric testing.
Quantification of bundling behavior of M. vaginatus
Differential bundling behavior of M. vaginatus was observed during co-culture trials. To gauge this effect specifically, co-cultures of cyanosphere isolates and M. vaginatus were repeated using the isolates previously described (Table 1) on nitrogen-replete (N+) or nitrogen-deplete (N−), carbon-free medium solid plates (10% BG11 and 10% BG110 [57], respectively) for each isolate tested (n = 3), allowed to grow for 14 days at 23 °C, 18–20 μE m−2 s−1, 14 h light cycle before being microscopically counted. Sixty bundles per treatment were chosen by dividing the three petri plates into quadrants and selecting five bundles at random in each quadrant. Bundles were selected along a straight transect of the quadrant to avoid duplication in counting. We determined the number of trichomes contained at three random cross-sections along the length of the bundle and calculated an average. Differences in average bundle size (number of trichomes per bundle) according to treatment were tested for significance using R [52] with a nonparametric Wilcox or Kruskal–Wallis tests, as appropriate.
Results
Cyanosphere community manipulation and nitrogen-fixation potential assessment
Cyanosphere community compositions differed even at the phylum level (Fig. S1A), with Bacteroidetes, Proteobacteria, non-Microcoleus Cyanobacteria, and Chloroflexi accounting for more than 75% of the reads in the N-addition and P-addition treatments and more than 50% of the reads in the controls. These differences were significant between control and either N-addition or P-addition treatments (PERMANOVA, P < 0.03), but no significant difference was detected between the N-addition and the P-addition treatments (PERMANOVA, P = 0.34) (Fig. S1B). At the sequence variant level, cyanosphere communities were significantly different in composition across all treatments (PERMANOVA, P < 0.001) (Fig. 1a). A SIMPER analysis showed that the P-addition treatment and the control were enriched in potential nitrogen-fixing taxa such as Azohydromonas, Bacteroides, Methylobacterium, Microvirga, and Ralstonia (Table S1A), over the N-addition treatment. Interestingly, the N-addition treatment resulted in a cyanosphere enriched in bacterial taxa such as Fluviicola, Burkholderia, Pseudoxanthomonas, and Achromobacter that are known for phosphate solubilization, EPS-production, and siderophore production ability (Table S1B).
Fig. 1. Comparison of bacterial composition and nitrogen-fixation potential of M. vaginatus cyanosphere communities with varying nutrient additions.
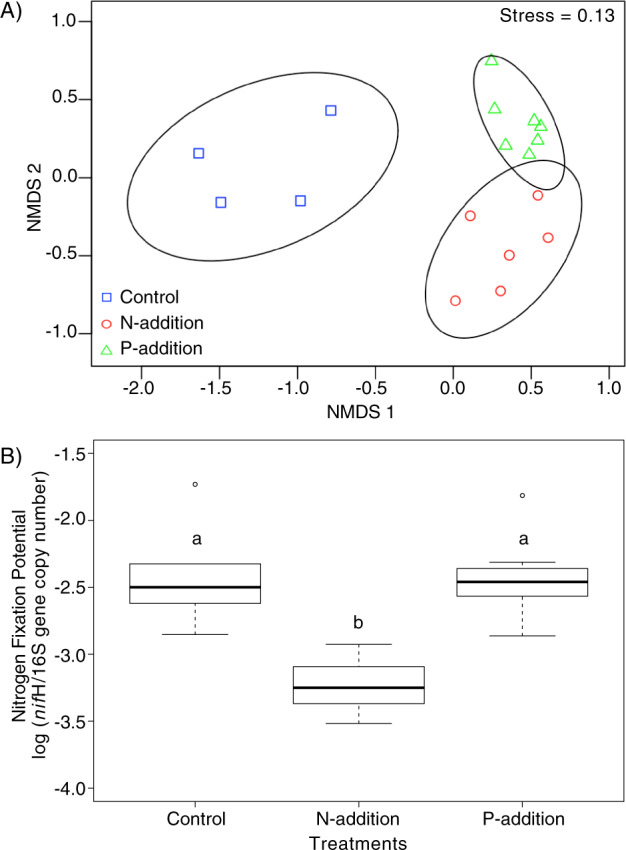
a NMDS ordination of community dissimilarity of M. vaginatus cyanospheres (excluding M. vaginatus itself) treated with nutrient additions (P-addition: n = 7, N-addition: n = 6) and untreated controls (n = 4), based on Bray–Curtis pairwise distances computed on the Hellinger-transformed sequence level composition with 95% confidence ellipses drawn for each, with a stress value of 0.13. Cyanosphere communities show significant differences among treatments and controls (PERMANOVA, P < 0.001). b Ratio of the nifH to 16S rRNA gene copy number in M. vaginatus cyanosphere communities in the same samples as in (a), showing significant differences between treatments (one-way ANOVA, P < 0.001), with control and P-addition treatment having significantly higher ratios than those in the N-addition treatment (Tukey, P < 0.002), but were not significantly different from each other (Tukey, P = 0.993).
Functionally, nifH/16S gene copy number ratios showed that the bundle cyanosphere communities differed significantly in nitrogen-fixation potential according to treatment (ANOVA, P < 0.001) (Fig. 1b), where N-addition resulted in significantly lower nitrogen-fixation potential (more than fivefold) over controls or P-addition (Tukey, P < 0.002 for both), which did not differ between them (Tukey, P = 0.993).
Pedigree of nitrogen-fixing heterotrophs and cyanosphere isolate co-culture
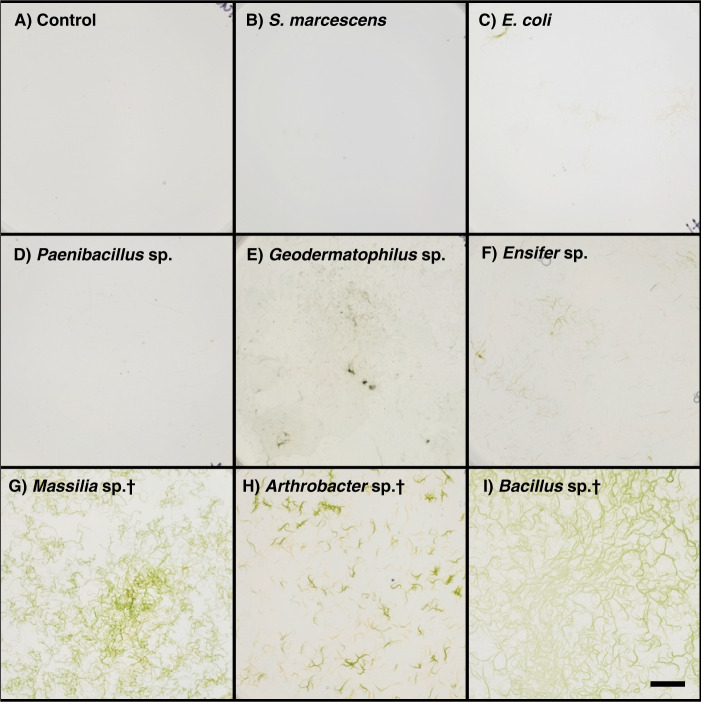
Nitrogen-free enrichments yielded 87 aerobic heterotrophic nitrogen-fixing isolates spanning several phyla. Most were Actinobacteria, Proteobacteria, and Firmicutes (Table S2). Although all isolates were screened for the presence of nifH genes, only 23 yielded positive amplification for the gene. Despite this, all isolates are maintained on nitrogen-free solid medium. Perhaps expectedly, only three isolates (3.4%) could be pedigreed to bona fide members of M. vaginatus cyanosphere communities by >98% 16S rRNA gene identity: Bacillus sp. O64, Arthrobacter sp. O80, and Massilia sp. METH4 (Tables 1, S2), which were chosen for experimentation. In co-culture, M. vaginatus PCC9802 showed a clear differential response to different heterotrophic isolates. Expectedly, it did not grow either alone or in co-culture with non-nitrogen-fixing bacteria on carbon-free, nitrogen-free solid medium (Figs. 2a–c, S2). It either grew slowly or failed to grow when co-cultured with nitrogen-fixing bacteria that are typical of biocrusts but are not associated with the cyanosphere (Figs. 2d–f, S2). However, it grew abundantly when co-cultured with the three nitrogen-fixing strains characteristic of the cyanosphere community (Figs. 2g–i, S2). Microscopic observations of co-cultures, particularly those with cyanosphere-associated heterotrophs, revealed copious growth of heterotrophic biomass in tight proximity to M. vaginatus bundles. Additionally, in separated growth trials, we confirmed that axenic M. vaginatus PCC9802 is unable to grow in nitrogen-free conditions (Fig. S3A) and all cyanosphere heterotrophs are unable to grow in carbon-free conditions (Fig. S3B).
Fig. 2. Co-cultures of M. vaginatus PCC9802 with heterotrophic bacteria on solid, carbon-, and nitrogen-free medium after 20 days of incubation (n = 3).
Scale bar is 1 cm. The heterotroph used varies by panel as: (a) no heterotrophs (control), (b) Serratia marcescens D1, (c) Eschrichia coli K12, (d) Paenibacillus sp. S14, (e) Geodermatophilus sp. B17, (f) Ensifer sp. S1, (g) Massilia sp. METH4, (h) Arthrobacter sp. O80, and (i) Bacillus sp. O64. Bacterial isolates matching cyanosphere members are denoted by (†).
Enhancement of M. vaginatus bundle-formation ability in culture
Incubation of axenic M. vaginatus cultures under nitrogen-deplete (N−) conditions resulted in a statistically significant (if moderate) increase in the average number of trichomes contained within bundles over that seen in incubations under nitrogen-replete medium (Figs. 3a, S4A). This resulted from a motility-based aggregation of trichomes rather than from growth, given that no net growth was observed on N deplete-medium (Fig. S3A). In additional tests, M. vaginatus was co-cultured with heterotrophs on N-free medium. Co-culture with two of three of the cyanosphere diazotrophs (Arthrobacter sp. O80 and Massilia sp. METH4, but not Bacillus sp. O64) resulted in statistically significant (Kruskal–Wallis test, P < 0.01; Figs. 3b, S4B) further enhancement of bundle formation over that measured in axenic M. vaginatus in the same medium. Co-culture with other bacteria (E. coli K12, Streptomyces sp. METH2, Ensifer sp. S1, S. marcescens D1, Geodermatophilus sp. B17, and Paenibacillus sp. S14) did not have the same effect or resulted in death of M. vaginatus PCC9802.
Fig. 3. Trichome content of axenic M. vaginatus bundles incubated under various conditions (n = 3).
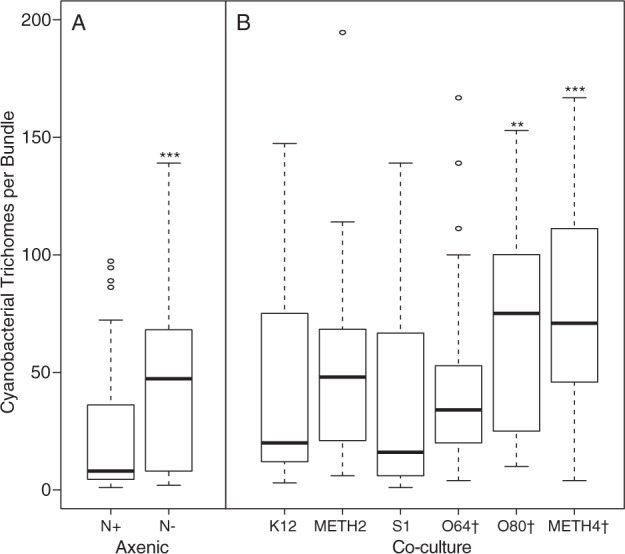
a Axenic M. vaginatus PCC9802 on nitrogen-deplete (N−) and nitrogen-replete (N+) solid medium. A Wilcox test shows significant differences (P < 0.001). b Co-cultured with various heterotrophic bacteria under N− conditions. Legend shows strain denomination and (†) denotes isolates matching known cyanosphere members. A Kruskal–Wallis test with chi-squared p value adjustments shows significant differences between axenic M. vaginatus on N− medium and co-cultures with isolates, Arthrobacter sp. O80 and Massilia sp. METH4 (P < 0.01 and <0.009, respectively), but not others. Co-cultures with S. marcescens D1, Geodermatophilus sp. B17, and Paenibacillus sp. S14 resulted in death of M. vaginatus and could not be quantified.
Discussion
Nutrient availability drives cyanosphere community composition
The addition of nutrients to cyanosphere communities caused a significant shift in their composition with respect to untreated controls, the shifts being distinct between treatments (Fig. 1a). This trend suggests that a relationship between M. vaginatus and the heterotrophic diazotrophs in the cyanosphere is dispensable in response to differential nutrient availability, thus implicating directly nutrient exchanges as the basis of a symbiosis. The fitness benefit under changing nutrient conditions would go, in this case, to the cyanobacterium who would continue to grow in a shifting environment by selecting appropriate new microbial partners only when needed. A similar behavior is a common occurrence in plant rhizosphere communities, where, depending on the needs of the plant, changes in exometabolite exudation profiles of plant roots can differentially attract or repel specific microbial constituents with specific functional traits [74, 75]. For the case of N, potentially diazotrophic members were washed out of the cyanosphere upon N-addition, but not upon P-addition. However, because taxonomy is only rarely a safe indicator of functional traits in bacteria, we sought confirmation through functional gene assessment. Again here, the relief of nitrogen limitation resulted in a cyanosphere community with a significantly lowered nitrogen-fixation potential (Fig. 1b) confirming the initial assessment. Interestingly, the addition of N resulted in an apparent enrichment in potentially P-solubilizing bacterial taxa, hinting to the notion that exchange of organic carbon for nutrients may go beyond C for N by using different heterotrophic partners. This must for now remain as a testable hypothesis.
M. vaginatus forms highly specific resource trading relationships under low nutrient availability
We were able to recreate the C for N resource trading relationship previously proposed [30] by reassembling its components in vitro through the use of pedigreed, heterotrophic bacteria isolated ad hoc from cyanosphere communities. We found that, despite belonging to different phyla, these heterotrophs typical of the cyanosphere community rapidly formed a mutualistic relationship with M. vaginatus in vitro allowing for copious growth of both partners under nitrogen-free, organic C-free conditions (Figs. 2g–i, S2), which would have allowed neither to thrive separately (Fig. S3). While not measured directly, the conspicuous growth of heterotrophic biomass was concentrated in tight proximity to M. vaginatus bundles (Fig. S4B), indicating that M. vaginatus was the source of organic carbon needed to complete the C for N mutualism. That this association is highly specific can be derived from the fact that co-culture with other heterotrophic diazotrophs commonly found in biocrusts (but not in the cyanosphere), failed to produce the same result (Figs. 2d–f, S2), as did co-culturing with heterotrophs from other environments (Figs. 2b, c, S2). Interestingly, these cooperative interactions seem to be absent or, in some cases, turn competitive when co-cultured on carbon- or nitrogen-replete medium, where no growth of M. vaginatus was observed (Fig. S5A).
Nitrogen limitation and mutualism induce bundling formation in M. vaginatus
Aggregation of M. vaginatus trichomes into macroscopic bundles enclosed by a common sheath is a morphogenetic trait for which the mechanism is unknown. In fact, it is the diacritical trait that defines the genus Microcoleus, although it is not expressed in pure culture [12]. While fitness benefits of this bundling behavior are clearly seen in an ability of various bundle formers to stabilize sedimentary substrates against erosion [19], it still presents potential drawbacks. Coming together into aggregates of large size will principally hamper uptake of diffusible nutrients, and cause self-shading [76], which is something that phototrophs avoid. Showing that the trait is inducible, as we did here, gives us a glimpse of what its fitness value might be based on. Our current observations implicate, by correlation, enhanced aggregation into bundles as a response to nitrogen limitation and, particularly also, to the establishment of mutualisms to thrive under such conditions. This aligns with observational evidence that the highly motile trichomes of M. vaginatus remain in their common sheath enclosure when pulled from environmental samples if kept under nitrogen limitation, but rapidly disperse from it when incubated in nitrogen rich medium (Fig. S5B). A priori, a dense supracellular structure with many trichomes competing for limited resources would not make sense, unless the resource would be a highly diffusible gas found in high concentration, like N2. Under nitrogen-fixing conditions, bundling should not present a major problem. But M. vaginatus also produced even larger bundles when co-cultured with nitrogen-fixing heterotrophs associated with cyanosphere communities, similar in size to those observed in field samples. The differential response of M. vaginatus to the presence of cyanosphere isolates suggest that increasing size may facilitate greater efficiency of resource trading, perhaps through the formation of more intense exometabolite gradients in another analogy to the rhizosphere of plants [75], or through the creation of localized oxygen-free micro-niches during the night, often measured within biocrusts [77] that might protect nitrogenase, as has been suggested for the planktonic cyanobacterium, Trichodesmium [78]. But the exact mechanisms remain to be tested with any rigor.
A mutualistic relationship of global reach and applied potential
Biocrusts communities are increasingly recognized as key not only to the local C and N biogeochemical cycles [79, 80], but also to its global dimension [81]. With a total biomass estimated at some 54 × 1012 g C [82], terrestrial cyanobacteria are one of the major components of biocrusts, and within them, M. vaginatus is the most common and widespread terrestrial cyanobacterium [17]. Given this background and the results presented here, it is likely that the mutualism of M. vaginatus with certain heterotrophs is responsible for a significant portion of terrestrial nitrogen-fixation at the global scale, and particularly important in the increasingly more expansive arid land areas suffering from loss of biocrust cover, be it by direct human impact [83, 84] or by effect of global change [18, 85], in which recovery hinges on the success of pioneer biocrusts formers. The results presented here also open the door to applied aspects of biocrust restoration and, in particular, to the sustainable production of biocrust inoculum for use in ecological restoration in dryland ecosystems. Current production approaches have focused solely on the phototrophic components [33, 34], but our results suggest that using selected heterotrophs to enhance production and survival of cyanobacterial inoculum may be a new avenue worth considering.
Conclusion
We provided strong evidence for a “carbon for nitrogen” resource trading relationship between M. vaginatus and certain members of its cyanosphere community by reconstructing the hypothesized mutualism from its components in the laboratory to gauge the ability of two-partner consortia to thrive in C- and N-free media and by showing that the relationship can be regulated by changing N availability. Further, we provide evidence that bundle formation by the cyanobacterium is tied to the conditions leading to mutualism, although its specific role remains to be discerned. We discuss the relevance of these findings for nitrogen cycling locally and globally, and their potential for translation to restoration ecology.
Supplementary information
Acknowledgements
We would like to thank Júlia Gomes Farias for her help with photography. This work was supported in part by the Jornada Basin LTER Graduate Research Fellowship Program and Center for Bio-mediated and Bio-inspired Geotechnics.
Author contributions
CN, AGS, and FGP conceived the research; CN, FGP designed experiments; CN performed experiments; CN analyzed data; CN, AGS, and FGP discussed results; CN, FGP wrote and edited the paper.
Data availability
Raw sequence data have been submitted to NCBI and are publicly available under BioProject PRJNA630480.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-020-00781-1) contains supplementary material, which is available to authorized users.
References
- 1.Garcia-Pichel F. Desert environments: biological soil crusts. encycl environ microbiol vol 6. New York, NY, USA: Set. Wiley-Interscience; 2003. pp. 1019–23. [Google Scholar]
- 2.Belnap J, Büdel B, Lange OL. Biological soil crusts: characteristics and distribution. biological soil crust: structure, function, and management. Berlin: Springer-Verlag; 2001. pp. 3–30. [Google Scholar]
- 3.Prăvălie R. Drylands extent and environmental issues. A global approach. Earth Sci Rev. 2016;161:259–78. [Google Scholar]
- 4.Garcia-Pichel F, Pringault O. Cyanobacteria track water in desert soils. Nature. 2001;413:380–1. doi: 10.1038/35096640. [DOI] [PubMed] [Google Scholar]
- 5.Pringault O, Garcia-Pichel F. Hydrotaxis of cyanobacteria in desert crusts. Microb Ecol. 2004;47:366–73. doi: 10.1007/s00248-002-0107-3. [DOI] [PubMed] [Google Scholar]
- 6.Soule T, Anderson IJ, Johnson SL, Bates ST, Garcia-Pichel F. Archaeal populations in biological soil crusts from arid lands in North America. Soil Biol Biochem. 2009;41:2069–74. [Google Scholar]
- 7.Nunes da Rocha U, Cadillo-Quiroz H, Karaoz U, Rajeev L, Klitgord N, Dunn S, et al. Isolation of a significant fraction of non-phototroph diversity from a desert biological soil crust. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu C, Zhang D, Huang Z, Liu Y. The vertical microdistribution of cyanobacteria and green algae within desert crusts and the development of the algal crusts. Plant Soil. 2003;257:97–111. [Google Scholar]
- 9.Bates ST, Nash TH, Garcia-Pichel F. Patterns of diversity for fungal assemblages of biological soil crusts from the southwestern United States. Mycologia. 2012;104:353–61. doi: 10.3852/11-232. [DOI] [PubMed] [Google Scholar]
- 10.Ullmann I, Büdel B. Ecological determinants of species composition of biological soil crusts on a landscape scale. In: Belnap J, Lange OL, editors. Biological soil crusts: structure, function, and management, 1st ed. Berlin: Springer-Verlag; 2001. pp. 203–13. [Google Scholar]
- 11.Lange OL, Belnap J, Reichenberger H, Meyer A. Photosynthesis of green algal soil crust lichens from arid lands in southern Utah, USA: role of water content on light and temperature responses of CO2 exchange. Flora. 1997;192:1–15. [Google Scholar]
- 12.Garcia-Pichel F, López-Cortés A, Nübel U. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microbiol. 2001;67:1902–10. doi: 10.1128/AEM.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Couradeau E, Karaoz U, Lim HC, Nunes Da Rocha U, Northen T, Brodie E, et al. Bacteria increase arid-land soil surface temperature through the production of sunscreens. Nat Commun. 2016;7:1–7. doi: 10.1038/ncomms10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeager C, Kornosky J, Housman DC, Grote EE, Belnap J, Kuske CR. Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado Plateau and Chihuahuan Desert. Appl Environ Microbiol. 2004;70:973–83. doi: 10.1128/AEM.70.2.973-983.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yeager CM, Kornosky JL, Morgan RE, Cain EC, Garcia-Pichel F, Housman DC, et al. Three distinct clades of cultured heterocystous cyanobacteria constitute the dominant N2-fixing members of biological soil crusts of the Colorado Plateau, USA. FEMS Microbiol Ecol. 2007;60:85–97. doi: 10.1111/j.1574-6941.2006.00265.x. [DOI] [PubMed] [Google Scholar]
- 16.Yeager CM, Kuske CR, Carney TD, Johnson SL, Ticknor LO, Belnap J. Response of biological soil crust diazotrophs to season, altered summer precipitation, and year-round increased temperature in an arid grassland of the Colorado Plateau, USA. Front Microbiol. 2012;3:1–14. doi: 10.3389/fmicb.2012.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Pichel F. Cyanobacteria. In: Schaechter M, editor. Encyclopedia of microbiology. 3rd ed. 2009. New York: Elsevier Inc.; 2009. p. 107–24.
- 18.Fernandes VMC, Machado de Lima NM, Roush D, Rudgers J, Collins SL, Garcia-Pichel F. Exposure to predicted precipitation patterns decreases population size and alters community structure of cyanobacteria in biological soil crusts from the Chihuahuan Desert. Environ Microbiol. 2018;20:259–69. doi: 10.1111/1462-2920.13983. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Pichel F, Wojciechowski MF. The evolution of a capacity to build supra-cellular ropes enabled filamentous cyanobacteria to colonize highly erodible substrates. PLoS ONE. 2009;4:4–9. doi: 10.1371/journal.pone.0007801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belnap J, Gardner J. Soil microstructure in soils of the Colorado Plateau: the role of the cyanobacterium Microcoleus vaginatus. West N Am Nat. 1993;53:40–7. [Google Scholar]
- 21.Starkenburg SR, Reitenga KG, Freitas T, Johnson S, Chain PSG, Garcia-Piche F, et al. Genome of the cyanobacterium Microcoleus vaginatus FGP-2, a photosynthetic ecosystem engineer of arid land soil biocrusts worldwide. J Bacteriol. 2011;193:4569–70. doi: 10.1128/JB.05138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajeev L, Nunes U, Klitgord N, Luning EG, Fortney J, Axen SD, et al. Dynamic cyanobacterial response to hydration and dehydration in a desert biological soil crust. ISME J. 2013;7:2178–91. doi: 10.1038/ismej.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper DU, Johnson L. Nitrogen limitation in dryland ecosystems: responses to geographical and temporal variation in precipitation. Biogeochemistry. 1999;46:247–93. [Google Scholar]
- 24.James JJ, Tiller RL, Richards JH. Multiple resources limit plant growth and function in a saline-alkaline desert community. J Ecol. 2005;93:113–26. [Google Scholar]
- 25.Neff JC, Reynolds RL, Belnap J, Lamothe P. Multi-decadal impacts of grazing on soil physical and biogeochemical properties in southeast Utah. Ecol Appl. 2005;15:87–95. [Google Scholar]
- 26.Schlesinger WH, Raikks JA, Hartley AE, Cross AF. On the spatial pattern of soil nutrients in desert ecosystems. Ecology. 1996;77:364–74. [Google Scholar]
- 27.Beraldi-Campesi H, Hartnett HE, Anbar A, Gordon GW, Garcia-Pichel F. Effect of biological soil crusts on soil elemental concentrations: Implications for biogeochemistry and as traceable biosignatures of ancient life on land. Geobiology. 2009;7:348–59. doi: 10.1111/j.1472-4669.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 28.Johnson SL, Budinoff CR, Belnap J, Garcia-pichel F. Relevance of ammonium oxidation within biological soil crust communities. Environ Microbiol. 2005;7:1–12. doi: 10.1111/j.1462-2920.2004.00649.x. [DOI] [PubMed] [Google Scholar]
- 29.Pepe-Ranney C, Koechli C, Potrafka R, Andam C, Eggleston E, Garcia-Pichel F, et al. Non-cyanobacterial diazotrophs mediate dinitrogen fixation in biological soil crusts during early crust formation. ISME J. 2016;10:287–98. doi: 10.1038/ismej.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couradeau E, Giraldo-Silva A, De Martini F, Garcia-Pichel F. Spatial segregation of the biological soil crust microbiome around its foundational cyanobacterium, Microcoleus vaginatus, and the formation of a nitrogen-fixing cyanosphere. Microbiome. 2019;7:111–22. doi: 10.1186/s40168-019-0661-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baran R, Brodie EL, Mayberry-lewis J, Hummel E, Nunes U, Rocha D, et al. Exometabolite niche partitioning among sympatric soil bacteria. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms9289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baran R, Ivanova N, Jose N, Garcia-Pichel F, Kyrpides N, Gugger M, et al. Functional genomics of novel secondary metabolites from diverse cyanobacteria using untargeted metabolomics. Mar Drugs. 2013;11:3617–31. doi: 10.3390/md11103617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velasco Ayuso S, Giraldo-Silva A, Nelson C, Barger NN, Garcia-pichel F. Microbial nursery production of high quality biological soil crust biomass for restoration of degraded dryland soils. Appl Environ Microbiol. 2017;83:1–16. doi: 10.1128/AEM.02179-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giraldo-Silva A, Nelson C, Barger N, Garcia-Pichel F. Nursing biocrusts: isolation, cultivation and fitness test of indigenous cyanobacteria. Restor Ecol. 2019;27:793–803. [Google Scholar]
- 35.Büdel B, Darienko T, Deutschewitz K, Dojani S, Friedl T, Mohr KI, et al. Southern african biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Microb Ecol. 2009;57:229–47. doi: 10.1007/s00248-008-9449-9. [DOI] [PubMed] [Google Scholar]
- 36.Ferreira D, Garcia-Pichel F. Mutational studies of putative biosynthetic genes for the cyanobacterial sunscreen scytonemin in Nostoc punctiforme ATCC 29133. Front Microbiol. 2016;7:1–10. doi: 10.3389/fmicb.2016.00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science. 2013;340:1574–77. doi: 10.1126/science.1236404. [DOI] [PubMed] [Google Scholar]
- 38.Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilbert JA, Meyer F, Jansson J, Gordon J, Pace N, Tiedje J, et al. The Earth Microbiome Project: meeting report of the ‘1 EMP meeting on sample selection and acquisition’ at Argonne National Laboratory October 6 2010. Stand Genom Sci. 2010;3:249–53. [DOI] [PMC free article] [PubMed]
- 40.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:1–10. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:242–5. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clarke KR, Gorley RN. PRIMER v6: user manual/tutorial. Prim Plymouth UK. 2006;7:192. [Google Scholar]
- 49.Nübel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–32. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ando S, Goto M, Meunchang S, Thongra-ar P, Fujiwara T, Hayashi H, et al. Detection of nifH Sequences in Sugarcane (Saccharum officinarum L.) and Pineapple (Ananas comosus [L.] Merr.) Soil Sci Plant Nutr. 2005;51:303–8. [Google Scholar]
- 51.Van Dorst J, Siciliano SD, Winsley T, Snape I, Ferrari BC. Bacterial targets as potential indicators of diesel fuel toxicity in subantarctic soils. Appl Environ Microbiol. 2014;80:4021–33. doi: 10.1128/AEM.03939-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- 53.Mirza BS, Rodrigues JLM. Development of a direct isolation procedure for free-living diazotrophs under controlled hypoxic conditions. Appl Environ Microbiol. 2012;78:5542–9. doi: 10.1128/AEM.00714-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Döbereiner J, Marriel IE, Nery M. Ecological distribution of Spirillum lipoferum Beijerinck. Can J Microbiol. 1976;22:1464–73. doi: 10.1139/m76-217. [DOI] [PubMed] [Google Scholar]
- 55.Dobereiner J, Urquiaga S, Boddey RM. Alternatives for nitrogen nutrition of crops in tropical agriculture. Fertil Res. 1995;42:339–46. [Google Scholar]
- 56.Wilson PW, Knight SG. Experiments in Bacterial Physiology, 3rded. Minneapolis, Minnesota: Burgess; 1952 p. 62.
- 57.Stanier RY, Kunisawa R, Mandel R. Purification and properties of unicellular blue-green algae (Order Chroococcales) Bacteriological Reviews. 1971;35:171–205. doi: 10.1128/br.35.2.171-205.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, et al. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) BMC Microbiol. 2008;8:1–8. doi: 10.1186/1471-2180-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;4:283–90. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 62.Ofek M, Hadar Y, Minz D. Ecology of root colonizing Massilia (Oxalobacteraceae) PLoS ONE. 2012;7:1–12. doi: 10.1371/journal.pone.0040117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Souza RSC, Okura VK, Armanhi JSL, Jorrín B, Lozano N, Da Silva MJ, et al. Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep. 2016;6:1–15. doi: 10.1038/srep28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones D, Keddie RM. The Genus Arthrobacter. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes: volume 3: archaea. bacteria: firmicutes, actinomycetes. New York, NY: Springer New York; 2006. pp. 945–60. [Google Scholar]
- 65.Baldani JI, Rouws L, Cruz LM, Olivares FL, Schmid M, Hartmann A. The family oxalobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: alphaproteobacteria and betaproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 919–74. [Google Scholar]
- 66.Mayilraj S, Stackebrandt E. The family paenibacillaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: firmicutes and tenericutes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 267–80. [Google Scholar]
- 67.Slepecky RA, Hemphill HE. The Genus Bacillus–Nonmedical. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E, editors. The prokaryotes: volume 4: bacteria: firmicutes, cyanobacteria. New York, NY: Springer US; 2006. pp. 530–62. [Google Scholar]
- 68.Carareto Alves LM, de Souza JAM, Varani A, de M, Lemos EG, de M. The family rhizobiaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: alphaproteobacteria and betaproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 419–37. [Google Scholar]
- 69.Normand P, Daffonchio D, Gtari M. The family geodermatophilaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: actinobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 361–79. [Google Scholar]
- 70.Kämpfer P, Glaeser SP, Parkes L, van Keulen G, Dyson P. The family streptomycetaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: actinobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 889–1010. [Google Scholar]
- 71.Octavia S, Lan R. The family enterobacteriaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The prokaryotes: gammaproteobacteria. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 225–86. [Google Scholar]
- 72.Poly F, Monrozier LJ, Bally R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol. 2001;152:95–103. doi: 10.1016/s0923-2508(00)01172-4. [DOI] [PubMed] [Google Scholar]
- 73.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, de Boer W, Garbeva P. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 2018;12:1252–62. doi: 10.1038/s41396-017-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhalnina K, Louie KB, Hao Z, Mansoori N, Da Rocha UN, Shi S, et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat Microbiol. 2018;3:470–80. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 76.Garcia-Pichel F. A model for internal self-shading in planktonic organisms and its implications for the usefulness of ultraviolet sunscreens. Limnol Oceanogr. 1994;39:1704–17. [Google Scholar]
- 77.Garcia-Pichel F, Belnap J. Small-scale environments and distribution of biological soil crusts. biological soil crusts: structure, function, and management. Berlin Heidelberg: Springer; 2001. pp. 193–201. [Google Scholar]
- 78.Paerl HW, Bebout BM. Direct measurement of O2-depleted microzones in marine oscillatoria: relation to N2 fixation. Science. 1988;241:442–5. doi: 10.1126/science.241.4864.442. [DOI] [PubMed] [Google Scholar]
- 79.Barger NN, Webber B, Garcia-Pichel F, Zaady E, Belnap J. Patterns and controls on nitrogen cycling of biological soil crusts. In: Weber B, Caldwell MM, Jayne B, Bettina W, Büdel B, Belnap J, et al. editors. Biological soil crusts: an organizing principle in drylands, 2nd ed. Switzerland: Springer; 2016. p. 257–85.
- 80.Sancho L, Belnap J, Colesie C, Raggio J. Carbon budgets of biological soil crusts at micro-, meso-, and global scales. In: Belnap J, Weber B, Burkhard B, editors. Biological soil crusts: an organizing principle in drylands. Switzerland: Springer; 2016. p. 287–304.
- 81.Weber B, Wu D, Tamm A, Ruckteschler N, Rodríguez-Caballero E, Steinkamp J, et al. Biological soil crusts accelerate the nitrogen cycle through large NO and HONO emissions in drylands. Proc Natl Acad Sci USA. 2015;112:15384–9. doi: 10.1073/pnas.1515818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Garcia-Pichel F, Belnap J, Neuer S, Schanz F. Estimates of global cyanobacterial biomass and its distribution. Arch Hydrobiol Suppl Algol Stud. 2003;109:213–27. [Google Scholar]
- 83.Belnap J, Eldridge D. Disturbance and recovery of biological soil crusts. In: Belnap J, Lange O, editors. Biological soil crusts: structure, function and management. Berlin: Springer; 2001. pp. 363–83. [Google Scholar]
- 84.Zaady E, Eldridge DJ, Bowker MA. Effect of local-scale disturbance on biocrusts. In: Weber B, Büdel B, Belnap J, editors. Biological soil crusts: an organizing principle in drylands. Cham: Springer International Publishing; 2016. pp. 429–50. [Google Scholar]
- 85.Williams WJ, Eldridge DJ, Alchin BM. Grazing and drought reduce cyanobacterial soil crusts in an Australian Acacia woodland. J Arid Environ. 2008;72:1064–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence data have been submitted to NCBI and are publicly available under BioProject PRJNA630480.