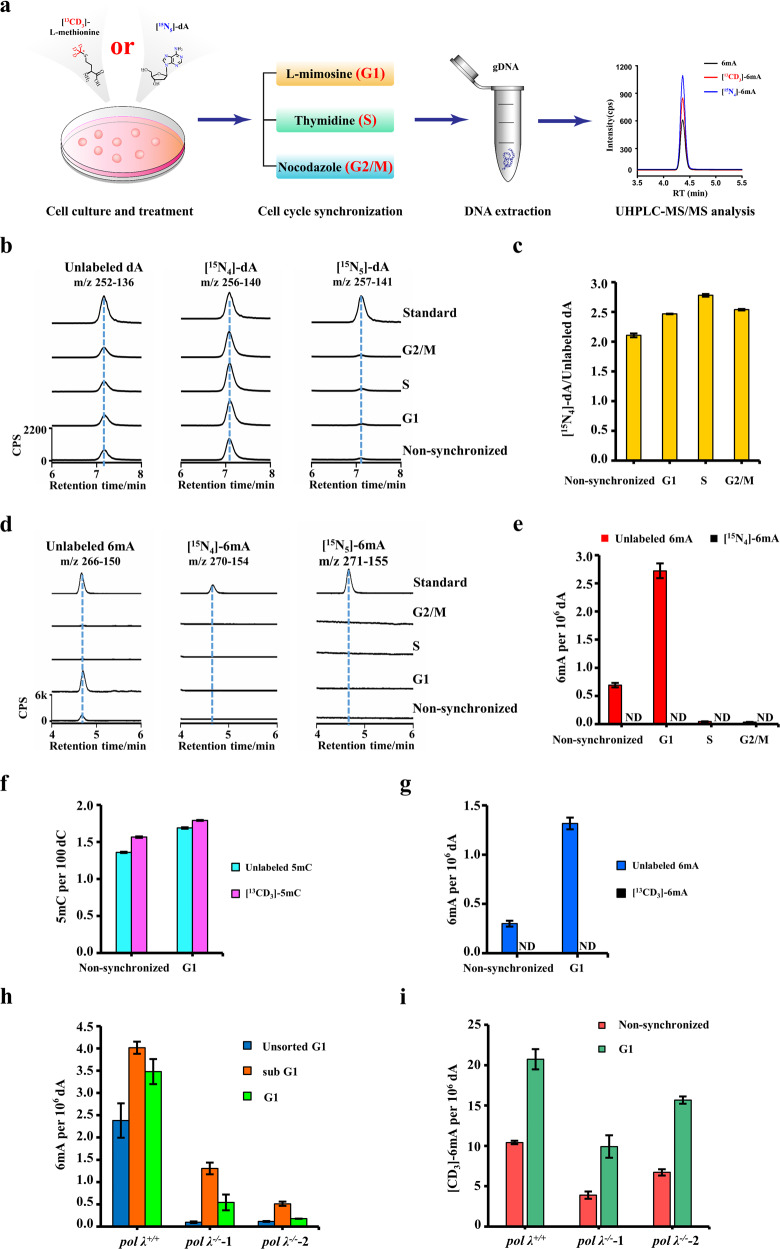
Fig. 1. Genomic incorporation of DNA N6-methyladenine and the contribution of DNA polymerase λ.
a Flow diagram of tracing DNA 6mA in mES cells by [15N5]-dA or [13CD3]-L-methionine. (b, c) UHPLC-MS/MS chromatograms (b) and quantification (c) of unlabeled dA, [15N4]-dA, and [15N5]-dA in the genome of mES cells. (d, e) UHPLC-MS/MS chromatograms (d) and quantification (e) of unlabeled 6mA, [15N4]-6mA, and [15N5]-6mA in the genome of mES cells. Note: [15N5]-dA was used as an initiation tracer and would be converted into [15N4]-dA in genomic DNA.9 The distinct cell cycle phases are indicated in (b–e). (f, g) UHPLC-MS/MS quantification of [13CD3]-5mC (f) and [13CD3]-6mA (g) in the genomes of non-synchronized and G1 phase ES cells. [13CD3]-L-methionine was used for tracing stable isotope-labeled methyl group. h UHPLC-MS/MS quantification of genomic 6mA levels in pol λ+/+ and pol λ−/− mES cells. The unsorted cells at G1 phase were directly obtained by L-mimosine treatment. The sorted cells at sub G1 and G1 phases were obtained by flow cytometry sorting of the L-mimosine-treated cells. i UHPLC-MS/MS quantification of the labeled genomic 6mA ([CD3]-6mA) levels in pol λ+/+ and pol λ−/− mES cells. The cells were treated with [CD3]-m6A alone or co-treated with [CD3]-m6A and L-mimosine. ND, not detected.