SUMMARY
Mature adipocytes store fatty acids and are a common component of tissue stroma. While adipocyte function in regulating bone marrow, skin, muscle, and mammary gland biology is emerging, the role of adipocyte-derived lipids in tissue homeostasis and repair is poorly understood. Here, we identify an essential role for adipocyte lipolysis in regulating inflammation and repair after injury in skin. Genetic mouse studies revealed that dermal adipocytes are necessary to initiate inflammation after injury and promote subsequent repair. We find through histological, ultrastructural, lipidomic, and genetic experiments in mice that adipocytes adjacent to skin injury initiate lipid release necessary for macrophage inflammation. Tamoxifen-inducible genetic lineage tracing of mature adipocytes and single cell RNA sequencing revealed that dermal adipocytes alter their fate and generate ECM-producing myofibroblasts within wounds. Thus, adipocytes regulate multiple aspects of repair and may be therapeutic for inflammatory diseases and defective wound healing associated with aging and diabetes.
Keywords: dermal adipose tissue, lipolysis, inflammation, wound healing, cellular plasticity, myofibroblast
Graphical Abstract
In Brief
Using genetic mouse models and transcriptomic profiling, Shook et al. show that skin resident adipocytes undergo lipolysis to promote efficient macrophage inflammation after injury. Lipolysis also induces adipocyte-derived cells to dedifferentiate and generate diverse myofibroblasts that to generate extracellular matrix in the wound bed.
INTRODUCTION
Tissue repair requires communication between multiple cell types to repair damaged tissue. Adipocytes are emerging as critical niche cells within multiple tissues including the skin (Festa et al., 2011; Plikus et al., 2017; 2008; Zhang et al., 2015; 2019b), bone marrow (Zhou et al., 2017a), and mammary gland (Wang et al., 2018; Zwick et al., 2018). While adipocytes secrete cytokines to regulate regeneration of hematopoietic stem cells after injury (Zhou et al., 2017a), and the antimicrobial peptide cathelicidin to combat bacterial infections (Zhang et al., 2015), the molecular function of adipocytes in tissue repair is not fully understood.
Adipocytes store and break down triglycerides to rapidly release fatty acids (FAs) to support metabolism of surrounding cells and tissues. Adipocyte lipolysis, or triglyceride hydrolyzation, starts with the activation of rate-limiting adipose triglyceride lipase (ATGL), which releases a non-esterified free FA and forms diacylglycerol (Eichmann et al., 2012; Frühbeck et al., 2014; Miyoshi et al., 2008). Subsequent lipases catalyze the hydrolysis of the remaining FAs. While injection of adipocyte-derived lipids can be immunostimulatory in the omentum (Tynan et al., 2014) and lipid-derived eicosanoids such as prostaglandins contribute to tissue repair (Nelson et al., 2013), whether adipocyte lipolysis occurs after injury and contributes to tissue repair is not known.
Skin is an excellent model to explore the mechanisms by which adipocytes control tissue repair. A large population of adipocytes reside in dermal white adipose tissue (DWAT) underneath the fibroblast-rich portion of the dermis (Driskell et al., 2014). During hair follicle growth, cold exposure and bacterial infections (Festa et al., 2011; Kasza et al., 2014; Zhang et al., 2015), dermal adipocytes fill with lipid in the form of triglycerides (Rosen and Spiegelman, 2006). Interestingly, expression of the dominant negative A-ZIP/F protein under the Fabp4 promoter (Reitman and Gavrilova, 2000) or use of PPARg inhibitors can reduce adipogenesis in mice and alter fibroblast repopulation in skin wounds (Schmidt and Horsley, 2013). Since adipogenic progenitors can form myofibroblasts during skin repair (Shook et al., 2018), it remains unclear whether the wound healing phenotype in A-ZIP/F mice is due to defective immature adipogenic precursor cells, mature adipocytes, or other phenotypes like diabetes that develop in these mice (Reitman and Gavrilova, 2000; Schmidt and Horsley, 2013).
Here, we identify a key role for mature adipocytes in tissue repair in the skin. Using genetic strategies to selectively ablate skin resident adipocytes, we find that dermal adipocytes are necessary for robust macrophage inflammation after injury and efficient repair of epithelial and dermal cells. Surprisingly, we find that prior to macrophage infiltration, mature adipocytes undergo lipolysis, releasing FAs into skin wounds. Abrogation of dermal adipocyte lipolysis via genetic deletion of Atgl in mice results in decreased FA content in wound beds, reduced numbers of inflammatory macrophages and delayed repair. Additionally, genetic lineage tracing of mature adipocytes in mouse wounds, paired with single cell RNA sequencing (scRNA-seq) and transcriptional profiling revealed that after lipolysis, adipocyte-derived cells generate myofibroblasts after injury. Our data reveal that adipocyte lipolysis plays a key role during tissue repair, allowing mature adipocytes to impact skin inflammation and generate extracellular matrix (ECM)-producing stromal cells.
RESULTS
Dermal adipocytes contribute to skin wound healing
To examine the role of dermal adipocytes in skin repair while avoiding systemic metabolic and diabetic phenotypes associated with systemic loss of adipocytes (Moitra et al., 1998; Rochford, 2014), we intradermally injected a low dose of diphtheria toxin into mice expressing an inducible diphtheria toxin receptor (iDTR) in adipocytes (AdipoqCre; mT/mG; iDTR mice) (Zwick et al., 2018). Dermal adipocytes were efficiently ablated 4 days after diphtheria toxin (DT) administration, while total animal weight and other adipocyte depots, like the gonadal white adipose depot (GWAT), remained unchanged (Figure 1A). Additionally, the numbers of immune cells, including dendritic cells, Langerhans cells, T cells and macrophages, was similar between AdipoqCre+; iDTR and AdipoqCre−; iDTR mice in uninjured skin (Figure S1A–D).
Figure 1. Dermal adipocytes contribute to skin wound healing.
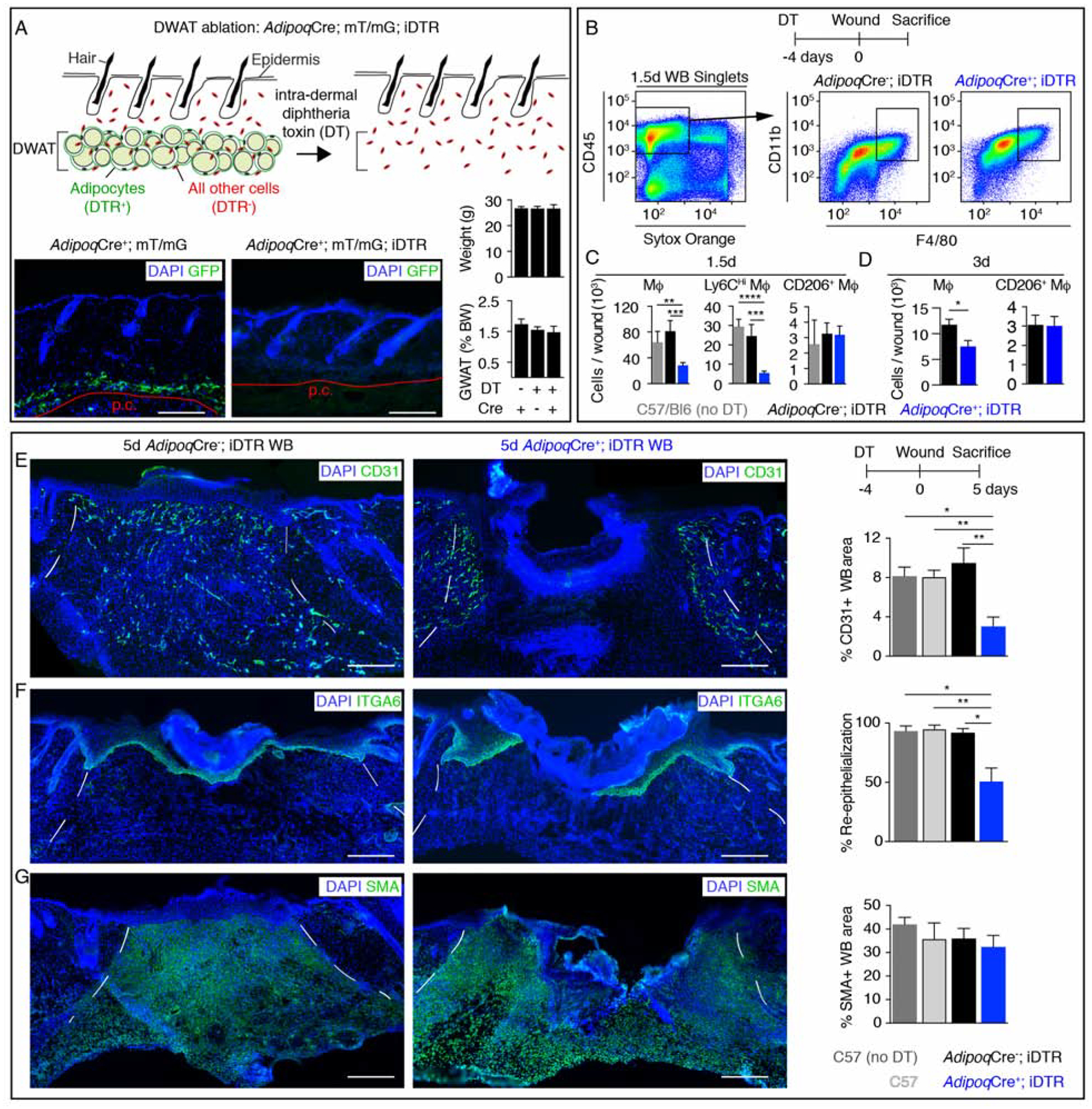
(A) Schematic and GFP immunostained images of uninjured skin and quantification of mouse body weight (BW) and the percentage of BW that is gonadal white adipose tissue (GWAT), following intradermal injection of diphtheria toxin (n ≥ 6 mice for each condition). Red line delineates the panniculus carnosus (p.c.). Scale bars, 250μm. (B–D) Flow cytometry plots (B) and quantification (C–D) of macrophages after injury (n ≥ 5 mice each condition and time point). (E–G) Immunostained sections and quantification of CD31 (revascularization) (E) (n ≥ 6 mice each condition), ITGA6 (re-epithelialization) (F) (n ≥ 6 mice each condition) and smooth muscle actin (SMA; fibroblast repopulation) (G) (n ≥ 4 mice each condition) in day 5 wound beds (WB). Scale bars, 250μm. Dotted white lines delineate wound edges. Error bars indicate mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001. DT, diphtheria toxin.
To examine whether wound repair was defective in skin lacking mature adipocytes, we examined the inflammatory phase of repair in AdipoqCre−; iDTR and AdipoqCre+; iDTR mice treated with diphtheria toxin (DT) prior to injury and C57/Bl6 mice that were not treated with DT. Ablating dermal adipocytes prior to injury resulted in a 65% reduction in macrophages in 1.5-day wound beds (Figure 1B and 1C, Figure S1A). Further, depletion of adipocytes did not alter the numbers or activation of other immune cells, in 1.5-day wound beds (Figure S1A–F). Specifically, wounds of mice lacking dermal adipocytes had fewer inflammatory Ly6CHi macrophages that are recruited during inflammation (Ramachandran et al., 2012; Willenborg et al., 2012), and not anti-inflammatory CD206+ macrophages (Figure 1B and 1C). Macrophages that did migrate into wound beds of adipocyte-depleted skin expressed similar levels of cytokine mRNAs as macrophages within adipocyte-containing skin (Figure S1G). Reduced numbers of wound bed macrophages persisted in wounds of adipocyte-depleted skin 3 days after injury (Figure 1D), when the local cytokine milieu transitions from inflammatory to anti-inflammatory and promotes repair (Crane et al., 2014; Shook et al., 2016).
We next examined skin repair by analyzing the proliferative phase of healing in DT-treated AdipoqCre−; iDTR, AdipoqCre+; iDTR, and wild-type mice, compared to untreated wild-type mice. While ablating macrophages throughout wound healing has been shown to impair all reparative processes (Clausen et al., 1999; Goren et al., 2010; Lucas et al., 2010; Mirza et al., 2010; Shook et al., 2016), ablating dermal adipocytes reduced revascularization by 66% throughout the entire wound bed and re-epithelialization by 50% in 5-day wound beds, with no detectable changes in SMA+ or ER−TR7+ fibroblast repopulation or wound contraction (Figure 1E–G, Figure S2A–G). Delayed revascularization and re-epithelialization in wounds from adipocyte-ablated skin was corrected 7 days after injury (Figure S2H–J), similar to when macrophages are ablated during the inflammation phase of wound healing (Lucas et al., 2010). These data suggest that dermal adipocytes are required for efficient activation of inflammatory responses that impact the efficiency of early stages of skin repair.
Dermal adipocytes undergo lipolysis after injury
Our adipocyte ablation data suggested that adipocytes impact repair within days after injury. Since adipocyte size and numbers increase rapidly in response to bacterial infection (Zhang et al., 2015), we examined the size of adipocyte lipid droplets and cell numbers at the periphery of wounds using perilipin 1 staining (PLIN1). Interestingly, we observed an initial increase in the average adipocyte area 16 hours after injury followed by a dramatic decrease in adipocyte size, while the number of PLIN1+ cells within 850μm from the wound edge did not change (Figure 2A). This decrease in adipocyte size temporally corresponded with infiltration of macrophages (Figure 2B) (Leibovich and Ross, 1975; Rodero et al., 2012; Shook et al., 2016).
Figure 2. Dermal adipocytes undergo lipolysis after injury.
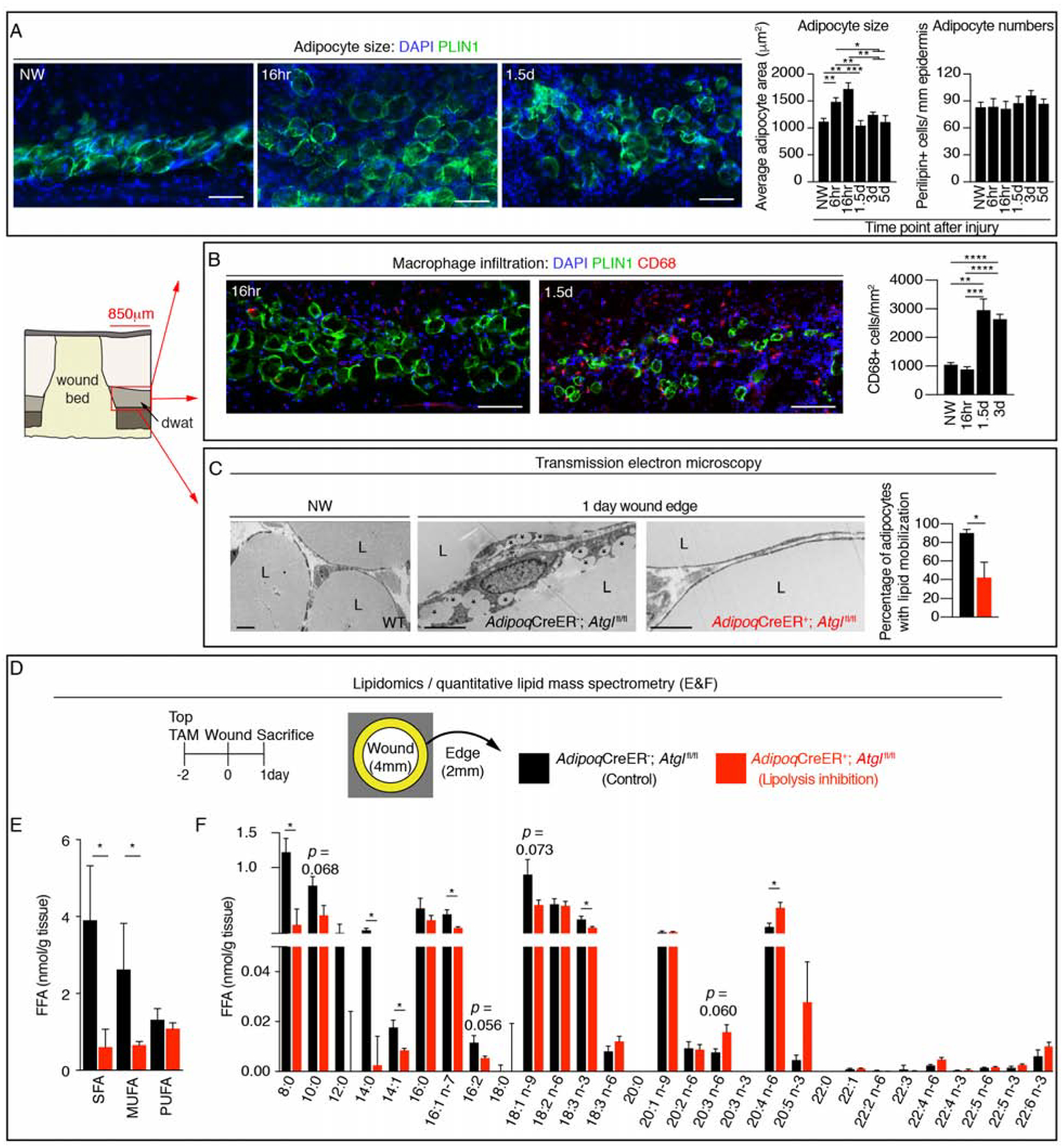
(A) Representative PLIN1 immunostained images of DWAT at the periphery of wounds and quantification of adipocyte cross sectional area and numbers (n ≥ 6 mice each time point). Scale bars, 100μm. (B) Representative immunostained images and quantification of CD68+ cells at the periphery of wound beds and in non-wounded (NW) skin (n ≥ 6 mice each time point). Scale bars, 100μm. (C) Transmission electron microscopy of adipocytes in uninjured skin and adipocytes at the wound periphery 1 day after injury. Asterisks show small lipid droplets. Quantification of adipocytes containing small lipid droplets at the periphery of a larger lipid droplet (n = 4 mice each condition). Scale bars, 5μm. (D) Experimental approach for quantitative lipidomics. (E–F) Lipid mass spectrometry quantification of non-esterified free fatty acid (FFA) classes (E) (n = 4–5 mice each condition) and FFA species (F) (n = 4–5 mice each time point). L, lipid droplet; SFA, saturated fatty acid; MUFA, monounsaturated fatty acid; PUFA, polyunsaturated fatty acid. Error bars indicate mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
See also Figure S3.
The reduction in adipocyte size at the wound edge suggested that adipocytes may be altering their lipid content through β-oxidation or lipolysis. To examine whether lipolysis of dermal adipocytes occurs after injury, we genetically targeted the first and rate-limiting enzyme in lipolysis, adipose triglyceride lipase (ATGL), to be deleted in adipocytes by crossing AdipoqCreER; mT/mG mice with Atglfl/fl mice. We optimized this system to preferentially target dermal adipocytes with a topical dose of tamoxifen, which resulted in Cre activity (defined by GFP+ cells) in ~60% of dermal PLIN 1+ cells while only ~20% of subcutaneous (SWAT) and gonadal white adipose tissue (GWAT) adipocytes were labeled (Figure S3A–E), without changing the SWAT or GWAT mass (Figure S3F). Since breakdown of the unilocular lipid droplet can result in the detection of multiple smaller droplets (Himms-Hagen et al., 2000; Miyoshi et al., 2008), we examined wound edge adipocytes in control and AdipoqCreER+; Atglfl/fl mice by transmission electron microscopy (Bani et al., 2013; Himms-Hagen et al., 2000; Varghese et al., 2019; Zwick et al., 2018). While wound edge adipocytes contained small lipid droplets in addition to a larger lipid droplet in AdipoqCreER−; Atglfl/fl mice, unilocular lipid droplets were retained in wound edge adipocytes in AdipoqCreER+; Atglfl/fl mice, similar to adipocytes in non-wounded skin (Figure 2C). Importantly, quantitative lipid mass spectrometry of non-esterified FAs revealed that genetic inhibition of dermal adipocyte lipolysis reduced saturated fatty acids (SFA) and monounsaturated fatty acids (MUFA) by 50% at the wound edge 24 hours after injury (Figure 2D–E). No difference was observed in the quantity of poly-unsaturated fatty acids (PUFA). Taken together, these data support that skin injury stimulates dermal adipocyte lipolysis to promote SFA and MUFA release into the site of injury.
Impairing adipocyte lipolysis reduces macrophage numbers during inflammation
To characterize dermal adipocyte FAs, we performed quantitative lipid mass spectrometry on purified adipocytes from DWAT and SWAT depots (Zwick et al., 2018). Interestingly, various long chain PUFA, including pro-inflammatory immune modulating arachidonic acid (20:4 n-6), are enriched in DWAT adipocytes compared to SWAT adipocytes (Figure S3G). Additionally, arachidonic acid was elevated during the inflammatory phase of wound healing (Figure S3H), which corresponds to the timing of dermal adipocyte lipolysis after injury.
To determine if a specific FA species was mobilized by injury-induced lipolysis, we performed quantitative lipid mass spectrometry on wounds derived from AdipoqCreER−; Atglfl/fl and AdipoqCreER+; Atglfl/fl mice. While we did not observe differences in free FA species present in skin of non-wounded AdipoqCreER−; Atglfl/fl and AdipoqCreER+; Atglfl/fl mice (Figure S3I–M), several FAs, including palmitoleic acid (16:1 n-7), oleic acid (18:1 n-9), and α-linoleic acid (18:3 n-3) were reduced in wound beds of AdipoqCreER+; Atglfl/fl mice compared to control wounds (Figure 2F). Interestingly, despite the contribution of prostaglandins to skin repair (Dhall et al., 2015; Futagami et al., 2002; Kendall and Nicolaou, 2013; Nelson et al., 2013; Nicolaou, 2013), no differences in eicosanoid derivatives were detected in AdipoqCreER+; Atglfl/fl mice compared to control mice at 24 hours post injury (Figure S3N–Q). Taken together, these data suggest that free FAs released by mature adipocytes proximal to the wound occurs within 24 hours after skin injury.
Since dermal adipocytes are required for efficient macrophage infiltration after injury (Figure 1B–D), and reduced adipocyte size and lipid dynamics occur concomitant with an increase in macrophages after injury (Figure 2A–B), we examined if adipocyte lipolysis contributes to inflammation in skin wounds. Genetic inhibition of dermal adipocyte lipolysis in AdipoqCreER+; Atglfl/fl mice did not alter the relative number of immune cells in uninjured skin (Figure S4A–B). However, inhibition of dermal adipocyte lipolysis reduced total and monocyte-derived Ly6CHi wound bed macrophages by ~50% 1.5 days after injury compared to control mice (Figure 3A–B), but did not alter other immune cells (Figure S4B). By 3 days after injury, the reduced number of wound bed macrophages was no longer detectable (Figure 3C), suggesting that the ~40% of adipocytes able to undergo lipolysis (Figure S3A) or non-FA-dependent signaling is sufficient to induce macrophage formation in wounds.
Figure 3. Impairing adipocyte lipolysis reduces macrophage numbers during inflammation.
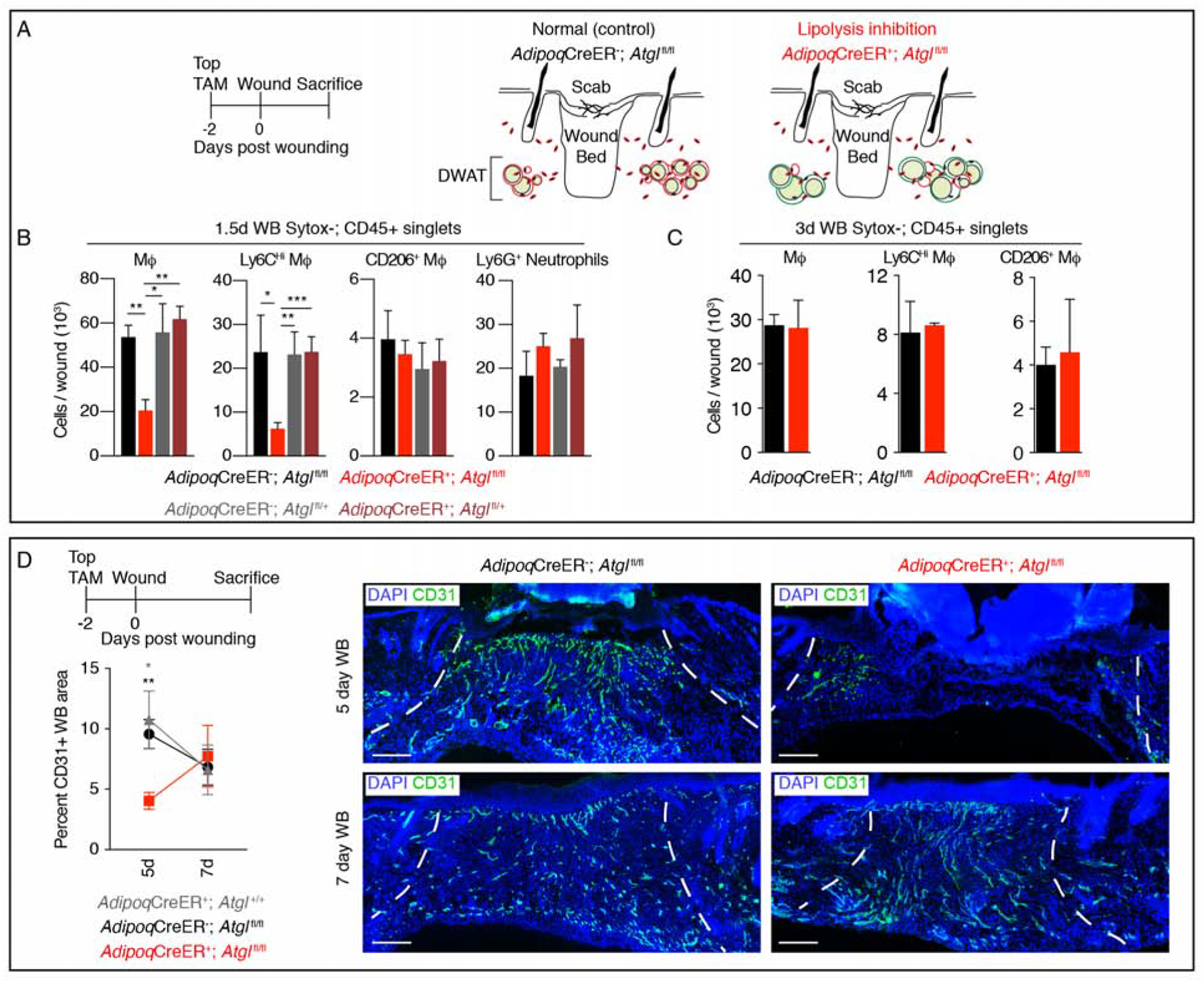
(A) Schematic of the strategy to inhibit lipolysis during wound healing. (B) Flow cytometry quantification of macrophages, macrophage subsets and neutrophils in 1.5-day wound beds (WB) (n = 5–7 mice each condition). (C) Flow cytometry quantification of macrophages and macrophage subsets in 3-day wound beds (n = 3–4 mice each condition). (D) CD31 immunostained sections and quantification 5 and 7 days after injury (n = 6 mice 5-day; n ≥ 3 mice 7-day). Scale bars, 250μm. Dotted lines indicate wound edges. Error bars indicate mean ± SEM. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
See also Figure S4.
To explore the impact of reduced dermal adipocyte lipolysis to the proliferative phase of repair, we analyzed re-vascularization, re-epithelialization, and fibroblast repopulation. While fibroblast repopulation and re-epithelialization were not significantly altered (Figure S4C–F), a ~50% reduction in revascularization was observed in day 5 wound beds that was corrected by 7 days after injury (Figure 3D, and Figure S4F). To determine if dermal adipocyte lipolysis contributes to revascularization after macrophages are recruited to the site of injury, we administered tamoxifen to AdipoqCreER+; Atglfl/fl mice 2 days after injury and analyzed revascularization in day 5 wounds. Interestingly, revascularization was not altered when lipolysis was inhibited 2 days after injury (Figure S4G), but was similar to control mice from our adipocyte ablation and lipolysis inhibition experiments (Figure 1E, and Figure 3D). These data suggest that reduced macrophage numbers in wound beds of AdipoqCreER+; Atglfl/fl mice likely cause delayed revascularization after injury (Ferrante et al., 2013; Lucas et al., 2010; Macedo et al., 2010).
Given that inhibition of dermal adipocyte lipolysis did not alter proliferation of monocytes or macrophages (Figure S4H), lipolysis likely promotes monocyte/macrophage recruitment or monocyte differentiation into macrophages. Future studies will explore the mechanisms by which adipocyte-derived FAs impact inflammation in skin wounds.
Adipocyte-derived cells migrate into wound beds but do not refill with lipids
Recent work in Drosophila demonstrated that fat body cells, the adipocyte equivalent in flies, actively migrate to epidermal wounds and contribute to repair (Franz et al., 2018). To examine if mature adipocytes contribute to later stages of skin repair, we performed genetic lineage tracing of adipocytes prior to injury using AdipoqCreER; mT/mG mice (Figure 4A–B). To label newly differentiated adipocytes, we induced Cre activity during the inflammatory and proliferative phases of repair. Surprisingly, when we labeled adipocytes immediately prior to injury, small GFP+ cells were present within 7-day wounds (Figure 4B). However, GFP+ cells were absent in wound beds when we administered tamoxifen during the inflammatory or proliferative phases of repair (Figure 4C–D). Adipocyte-derived cells can be lineage traced into wounds using a single low dose of topical tamoxifen (Figure 4E). To further evaluate how adipocytes contribute to the regenerating dermis, we analyzed whether lipolysis was required for GFP+ cells to migrate into wounds. Indeed, Adipoq-traced cells were not detected in wound beds from mice with impaired dermal adipocyte lipolysis (Figure 4F). To determine if the lack of GFP+ cells resulted from an inability of tamoxifen to enter wound beds prior to revascularization, we treated Rosa26CreER; mT/mG mice with tamoxifen after injury and detected many GFP+ cells in wound beds (Figure 4G). These data indicate that lipid stores are lost in mature adipocytes after skin injury, allowing the cells to migrate into wound beds.
Figure 4. Adipocyte-derived cells migrate into wound beds.
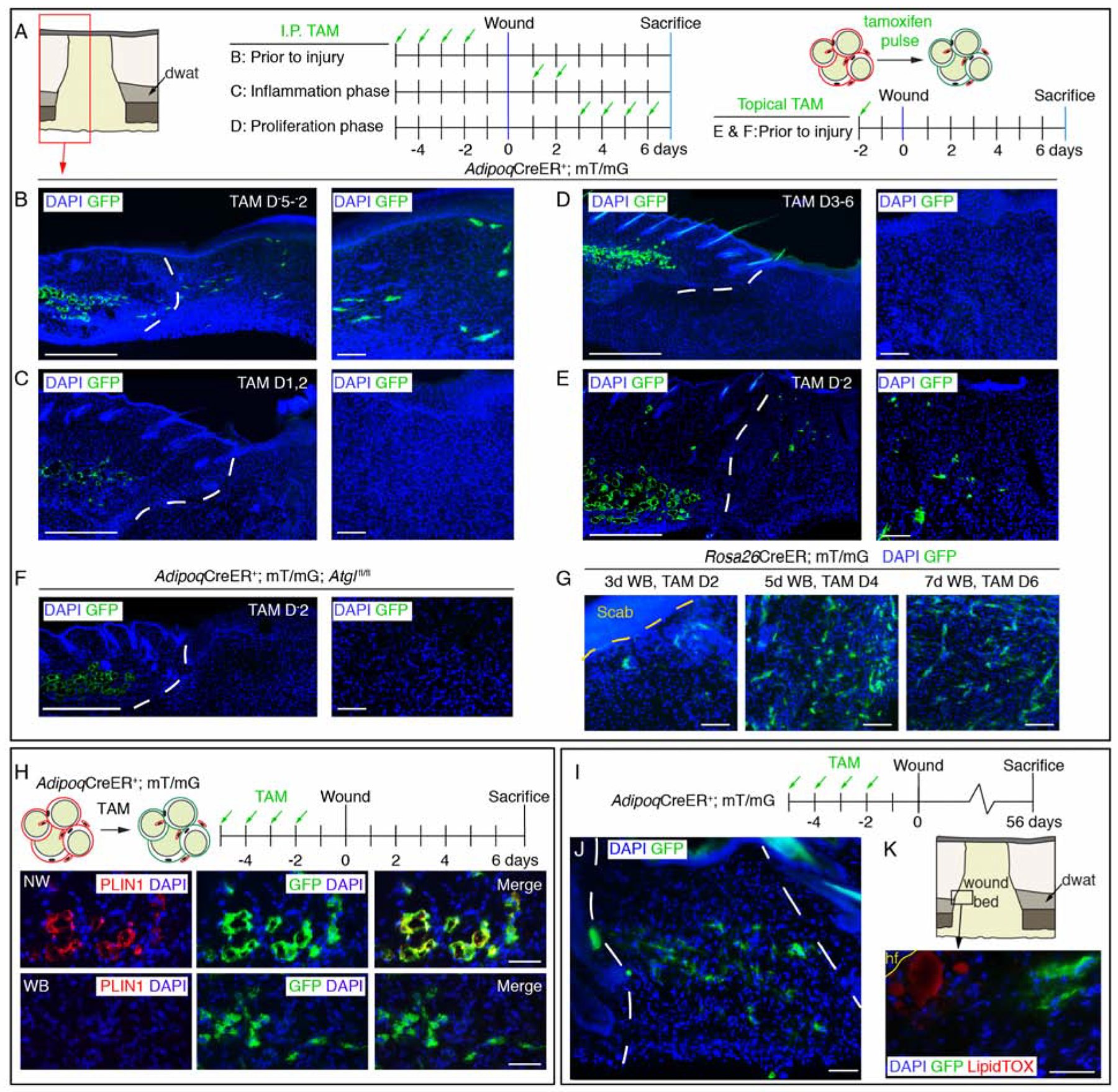
(A–F) Schematic of tamoxifen labeling strategy and immunostained images of GFP+ cells in day 7 wounds. Lower magnification images show dermal adipocytes (GFP+) and wound beds. Higher magnification panels are representative images from inside wound beds. Red box in A is representative of the wound bed location displayed in low magnification images in B–F. Scale bars, 500μm in lower magnification panels and 100μm in higher magnification panels. (G) Images of GFP+ cells in the center of wound beds from Rosa26CreER; mT/mG mice treated with tamoxifen at different stages of wound healing. (H) Schematic of adipocyte lineage tracing and PLIN1 immunostained sections in non-wounded (NW) skin and inside 7-day wound beds (WB). Scale bars, 100μm. (I) Labeling scheme to identify AdipoqCreER-traced cells. (J) Images of GFP+ cells in wound beds 8 weeks after injury. (K) LipidTOX staining at the wound edge near a growing peripheral hair follicle (hf). Scale bars, 50μm. White dotted lines delineate wound edges.
See also Figure S5.
To determine if the Adipoq promoter is active in other cell types prior to injury, we analyzed GFP+ expression in AdipoqCre+; mT/mG mice and AdipoqCreER; mT/mG mice using flow cytometry. While adipocytes in tissue sections were GFP+, we did not detect GFP+ cells in uninjured skin of AdipoqCreER+; mT/mG mice using flow cytometry (Figure S5A). However, small Pdgfra+, GFP+ cells existed within the stroma of AdipoqCre+; mT/mG mice (Figure S5B), consistent with the work of Zhang and colleagues (Zhang et al., 2019a). GFP+ stromal cells in AdipoqCre+; mT/mG mice expressed Cebpa, Pparg, Plin1, and Adipoq compared to the stromal vascular tissue fraction, consistent with adipocyte lineage identity (Figure S5C). Additionally, GFP+ cells did not colocalize with immune or endothelial cell markers (Figure S5D) or incorporate thymidine analogues administered before injury (Figure S5E, n = 7 wound beds, 0/1538 GFP+ cells were EdU+). Taken together, the contribution of GFP+ cells to the regenerating stroma of AdipoqCreER; mT/mG mice occurs from mature adipocytes that undergo lipolysis.
To further analyze the identify of Adipoq-traced cells in wound beds, we examined if GFP+ cells contained lipid droplets by immunostaining tissue sections with PLIN1. While all GFP+ cells colocalize with PLIN1 in uninjured AdipoqCreER+; mT/mG skin (Figure 4H) (Rivera-Gonzalez et al., 2017), we did not detect PLIN1 colocalization with GFP+ cells in day 7 wounds from AdipoqCreER+; mT/mG mice (Figure 4H). We next examined if Adipoq-traced cells were long-lived and persist after repair was complete. We observed many GFP+ cells in 6 of 8 wound beds 8 weeks after injury (Figure 4I–J). Surprisingly, these cells did not label with a neutral lipid dye, even though neighboring lipid-incorporating cells could be detected near growing hair follicles (Figure 4K) (Plikus et al., 2017). While GFP+ cells in uninjured skin were PLIN1+ (Figure 4H), GFP+ cells at the wound edge did not colocalize with PLIN1 or the transcription factor PPARg (Figure 5D) (Itabe et al., 2017; Rosen and Spiegelman, 2000). These data indicate that adipocyte-derived cells in wound beds are long-lived, but do not refill with lipids.
Figure 5. Adipocyte-derived cells in wound beds are long-lived and do not colocalize with adipocyte markers.
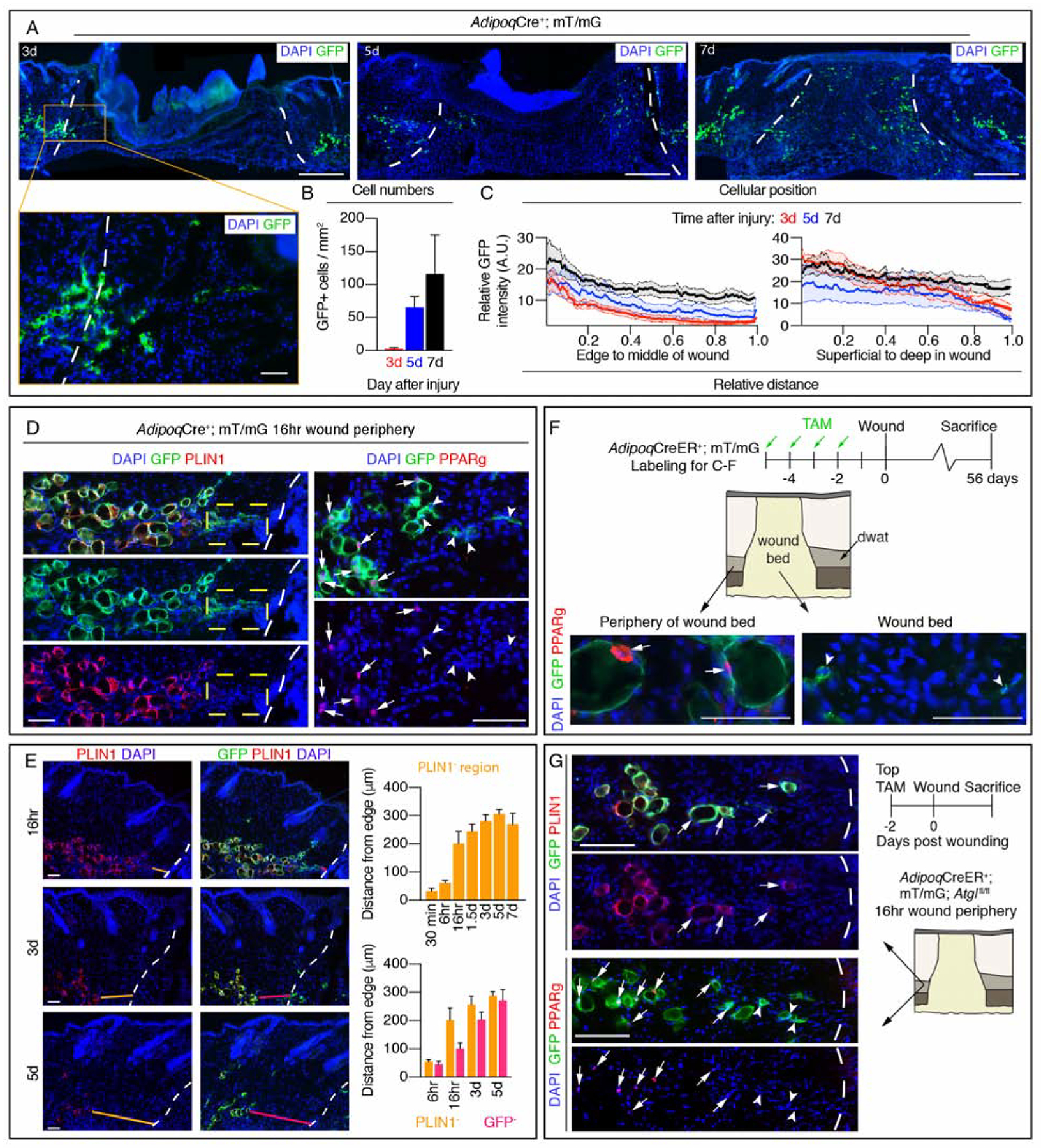
(A) Images of GFP in sections from the center of wound beds at different time points after injury. Scale bars, 500μm in lower magnification and 100μm in higher magnification (designated by orange box). (B) Quantification of GFP+ cells in wound beds (n = 4 mice each time point) and (C) the spatial distribution of GFP signal (n ≥ 7 mice each time point). (D) Colocalization of PLIN1 or PPARg with GFP+ adipocytes at the periphery of wounds 16-hours after injury. Dotted box indicates an area of GFP+ cells that lack PLIN1. Arrows indicate GFP+; PPARg+ cells, arrow heads indicate GFP+; PPARg− cells. Scale bars, 100μm. (E) PLIN1 and GFP immunostained images of DWAT and quantification of regions devoid of PLIN1 (orange lines) or GFP (magenta lines) at the periphery of wound beds at different time points after injury (n ≥ 6 mice each time point). Scale bars, 100μm. (F) Labeling scheme to identify long-lived AdipoqCreER-traced cells and PPARg staining. Arrows indicate GFP+; PPARg+ cells, arrow heads indicate GFP+; PPARg− cells. Scale bars, 50μm. (G) Immunostaining of PLIN1 or PPARg with GFP in wound periphery adipocytes from AdipoqCreER+; mT/mG; Atglfl/fl mice 16 hours after injury. Arrows indicate GFP+; PLIN1+ or GFP+; PPARg+ cells, arrow heads indicate GFP+; PPARg− cells. Scale bars, 200μm. White dotted lines delineate the wound edge. A.U., arbitrary units. Error bars indicate mean ± SEM.
See also Figure S5.
The cellular composition of wound beds varies based on age and the size of the original wound (Guerrero-Juarez et al., 2019; Ito et al., 2007; Plikus et al., 2017; Schmidt and Horsley, 2013; Shook et al., 2018; Telerman et al., 2017). To examine the contribution of adipocyte-derived cells in different wound contexts, we injured AdipoqCre; mT/mG mice with 2, 4, 6, or 8mm biopsy punches and examined the numbers and distribution of GFP+ cells in 7-day wound beds. While many GFP+ cells were detected through the entire wound bed from 2mm wounds, GFP+ cells were less frequent and biased towards the edges and superficial regions of larger wounds (Figure S5F). These data suggest that Adipoq-traced cells do not populate wound beds in larger wound models that promote hair follicle and adipocyte formation (Ito et al., 2007; Plikus et al., 2017).
We next examined the timing and location of Adipoq-traced cells within the regenerating dermis and observed GFP+ cells inside the wound edge, next to the DWAT, within 3 days after injury (Figure 5A). The number of GFP+ cells in wound beds increased during the proliferative phase (Figure 5B), representing ~1.5 ± 0.4% of the total mesenchymal fraction by day 5. These cells were biased toward the superficial and wound edge sides of the wounds (Figure 5C), similar to the trajectory that cells follow from the upper dermis (Driskell et al., 2013). GFP+ cells lacking PLIN1 appeared within hours after injury within ~200μm from the wound edge (Figure 5E). Even 56 days after injury, Adipoq-traced cells in wounds do not colocalize with PPARg (Figure 5F), suggesting a stable loss of multiple mature adipocyte markers after injury.
Since migration of Adipoq-traced cells into wound beds is dependent on the lipolytic enzyme Atgl (Figure 4F) and Adipoq-traced cells lack PLIN1 and PPARg expression prior to migrating into wound beds (Figure 5D), we investigated if the lack of PLIN1 and PPARg was Atgl-dependent. Interestingly, after injury, adipocytes at wound edges in AdipoqCreER+; mT/mG; Atglfl/fl mice retained PLIN1+ lipid droplets, yet lacked PPARg immunoreactivity 16 hours after injury (Figure 5G). Retention of PLIN1 was also observed in AdipoqCreER+; mT/mG; Atglfl/+ mice 16 hours after injury (Figure S5G); however, GFP+ cells that lack PLIN1 immunoreactivity were observed at the wound edge in day 5 wound beds of AdipoqCreER+; mT/mG; Atglfl/+ mice (Figure S5G), but not AdipoqCreER+; mT/mG; Atglfl/fl mice (Figure S5H). These data indicate that wound edge adipocytes lose adipocyte-specific markers shortly after injury and require Atgl-mediated lipid breakdown to migrate into wound beds.
While DWAT adipocytes downregulate lipid associated markers and populate wound beds, adipocytes in the underlying fascia (Merrick et al., 2019) were also labeled in our topical tamoxifen protocol (Figure S5I–J). Although adipocytes in the superficial fascia could contribute to our findings, these cells are not abundant, suggesting that dermal adipocytes are the major contributor to our findings.
Adipocyte-derived cells become wound bed myofibroblasts after injury
To investigate the cellular identity of Adipoq-traced cells in wound beds, we compared the gene expression profile of Adipoq-traced cells within wound beds to myofibroblasts and mature adipocytes. Myofibroblasts were isolated based on the lack of the adipocyte precursor marker SCA1 and the presence of the profibrotic marker CD26 (Festa et al., 2011; Rinkevich et al., 2015; Shook et al., 2018). Mature adipocytes were isolated from both wound edges and uninjured skin. Principle component analysis of the transcriptional profiles revealed that Adipoq-traced cells were more similar to CD26+ myofibroblasts than mature adipocytes (Figure 6A). A deeper analysis of wound-associated samples revealed that Adipoq-traced cells were enriched for genes associated with myofibroblasts and wound healing (Figure 6B–E and Figure S6A–D), including α-smooth muscle actin (SMA, Acta2) and multiple collagens (Figure 6C–E).
Figure 6. Adiponectin traced cells gain a myofibroblast gene expression profile during skin repair.
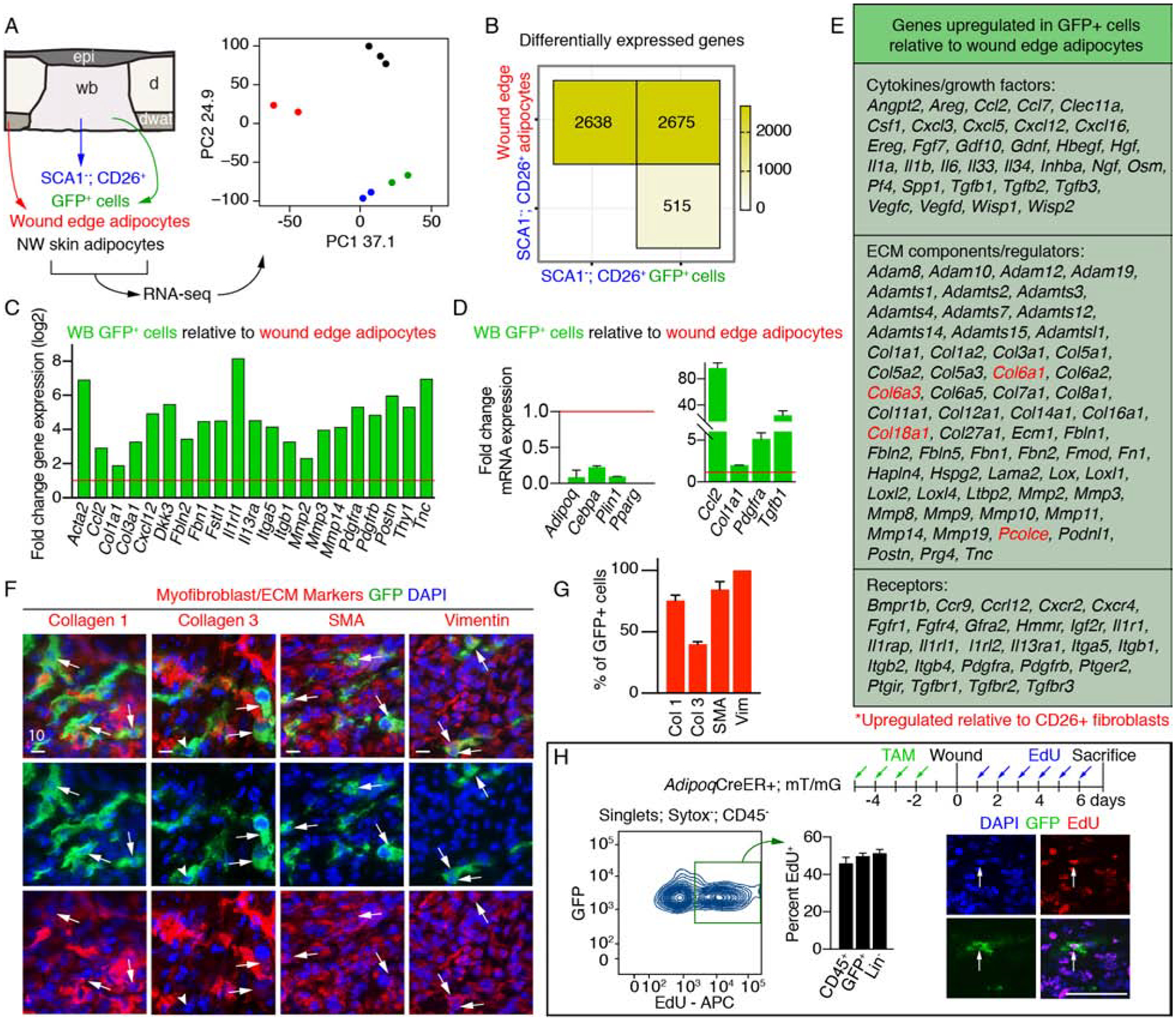
(A) Schematic and principal component (PC) analysis of cell populations isolated for RNA-seq. (B) Number of differentially expressed genes between cellular subsets. (C) Fold change in gene expression of myofibroblast-associated genes in GFP+ cells relative to wound edge/periphery adipocytes. Data from RNA-seq. (D) Quantitative real-time PCR of adipocyte signature genes (left) and myofibroblast genes (right) in GFP+ cells relative to wound periphery adipocytes (n = 3 mice). (E) Genes upregulated in GFP+ cells relative to wound edge adipocytes. Genes in red are also enriched compared to CD26+ myofibroblasts. (F–G) Images from day 5 wounds of AdipoqCre+; mT/mG mice (F) and quantification (G) of GFP+ cell colocalization with myofibroblast-associated markers (n = 3–5 mice). Scale bars, 10μm. (H) Flow cytometry plot and quantification of EdU+ cells (n = 3 mice). Scale bar, 100μm. Error bars indicate mean ± SEM. Epi, epidermis; d, dermis; dwat, dermal white adipose tissue; WB, wound bed.
See also Figure S6.
Interestingly, Ingenuity Pathway Analysis of transcriptional profiles from Adipoq-traced cells and mature adipocytes predicted that signaling pathways implicated in inflammation (IL1a and Il1b), fibrosis (Tgfβ1) and fibroblast proliferation (Tnf) (Barrientos et al., 2008; Werner and Grose, 2003) could account for differentially expressed genes between these populations (Figure S6B). Many of the biological functions predicted to be influenced by the gene expression profile of GFP+ cells were related to wound healing and connective tissue formation (Figure S6D), implicating adipocyte-derived cells as active participants in tissue formation after injury. The gene expression profile of adipocyte-derived cells was enriched for several cytokines and ECM remodeling molecules compared to wound edge dermal adipocytes (Figure 6E). Indeed, Adipoq-traced cells in the DWAT are SMA negative, even 3 days after injury (Figure S6E); however, GFP+ cells in wound beds express SMA (Acta2), Collagen 1, Collagen 3, Fibrillin, Fibronectin and Vimentin (Figure 6F–G and Figure S6F), and can migrate out of cultured wound bed explants (Figure S6G).
Since the transcriptome of Adipoq-traced cells revealed altered gene expression of transcription factors and enzymes that regulate cell cycle exit in adipocytes, such as Cdk4, Ccnd1, Cebpa, Klf2, Pparg (Farmer, 2006), we examined if these cells re-enter the cell cycle and proliferate. Indeed, almost 50% of GFP+ cells in wound beds incorporated EdU after injury, which is similar to the percent of proliferative immune cells (CD45+) and mesenchymal cells (CD45− and CD31−: Lin−) within wounds (Figure 6H). These data demonstrate that adipocyte-derived cells re-enter the cell cycle in response to wound-induced signaling.
We recently reported that several subsets of myofibroblasts exist in skin wound beds (Shook et al., 2018). Compared to the published transcriptomes of these myofibroblast subsets (Shook et al., 2018), Adipoq-traced cells have a distinct gene expression profile (Figure S6H). In the mammary gland during lactation and during hair cycling, mature adipocytes were shown to form adipocyte precursors (AP) (Wang et al., 2018; Zhang et al., 2019a). In contrast, Adipoq-traced cells in skin wounds remained negative for AP molecular markers CD26, CD34 and SCA1 (Figure S6I) (Festa et al., 2011; Rivera-Gonzalez et al., 2016; Rodeheffer et al., 2008; Shook et al., 2018; Zhang et al., 2019a). Interestingly, the gene expression profile of Adipoq-traced cells was enriched for many wound-associated secreted factors and expressed several distinct ECM molecules (Figure S6J) compared to other myofibroblasts. Unlike APs that become myofibroblasts during repair (Shook et al., 2018), Adipoq-traced cells are not enriched for genes associated with collagen maturation and crosslinking (Figure S6J), supporting that adipocyte-derived myofibroblasts have a distinct gene expression profile, and possibly a unique function.
To further explore how Adipoq-traced myofibroblasts compare to other myofibroblast subpopulations, we FACS purified lineage-negative (CD45− and CD31−) cells and performed scRNA-seq on wound beds and uninjured skin from AdipoqCre; mT/mG mice. Unsupervised clustering revealed 10 fibroblast subpopulations (Figure 7A and B, and Figure S7, A–C). Four fibroblast populations were enriched in wound bed samples compared to uninjured skin (FB2, FB4, FB5 and FB6) (Figure 7B). These clusters were enriched for myofibroblast markers and ECM-molecules associated with wound healing (Figure 7B), and had similar differential expression of genes previously described (Guerrero-Juarez et al., 2019). In support of our bulk RNA-seq and tissue immunostaining data, GFP-expressing cells from wound beds were enriched in clusters that expressed Acta2, Col1a1, Col3a1, Crip1, Pdgfra, Tnc and Vimentin (Figure 7C and Figure S7D–F).
Figure 7. Dermal adipocyte-derived cells become myofibroblasts after injury.
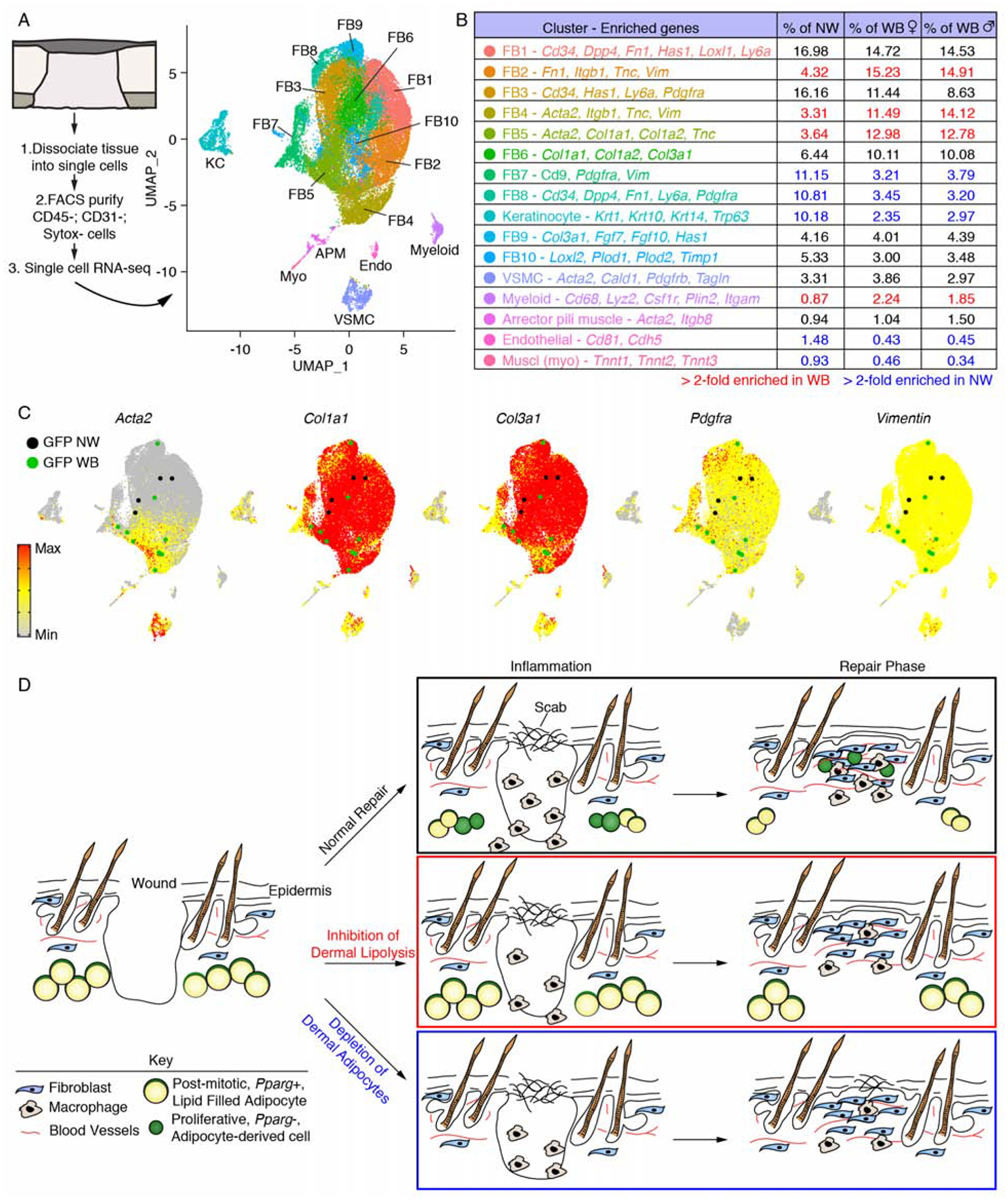
(A) scRNA-seq was performed on non-wounded (NW) skin and day 5 wound beds (WB) from AdipoqCre; mT/mG mice. UMAP dimension reduction plot is displayed with a table (B) identifying cell clusters based on enriched genes and quantifying their relative abundance. (C) Gene expression plots showing distribution of Acta2, Col1a1, Col3a1, Pdgfra and Vimentin. Expression levels for each cell are shown as Pearson residuals and displayed using a color scale, overlaid onto the UMAP plot. GFP+ cells from NW and WB samples are overlaid to emphasize their location. (D) Schematic illustration showing the contribution of adipocytes to skin wound healing. Following injury, adipocytes undergo lipolysis that supports macrophage inflammation. Adipocytes depleted of lipids become myofibroblasts and proliferate. Depletion of dermal adipocytes and inhibition of dermal adipocyte lipolysis reduces macrophage numbers during inflammation and delays revascularization. Ablating dermal adipocytes also delays re-epithelialization. FB, fibroblast; VSMC, vascular smooth muscle cell.
To better understand the transcriptional differences that define these populations, we analyzed genes upregulated in wound-enriched fibroblast clusters by comparing FB2, FB4 and FB5 to FB6, which had the lowest Acta2 expression among wound-associated fibroblasts (Figure S7G). Each subset had distinct genes and biofunctions associated with its gene expression profile (Table S1 and S2), suggesting they may be functionally unique myofibroblast populations. We compared myofibroblast-associated gene expression in Adipoq-traced cells to wound-associated fibroblast populations and used keratinocytes and macrophages/myeloid cells as reference cells (Figure S7H). In line with our bulk RNA-seq data (Figure 6A–E), GFP-expressing cells from wound beds had a distinct expression pattern from GFP-expressing cells in uninjured skin. GFP+ cells from wound beds lacked enrichment of AP markers Cd34 and Ly6a and appeared more similar to FB4 and FB5 in expression of many ECM-related genes and secreted molecules (Figure S7H). Additional experiments exploring the heterogeneity of Adipoq-derived myofibroblasts may reveal functional differences among these cells in skin wound beds.
Similar to a recent report (Zhang et al., 2019b), GFP+ cells, from both uninjured skin and wound beds, that remained intact and passed our quality control criteria did not have detectable levels of Adipoq, Lep, Plin1 or Pparg. Given that we detect these genes in FACS-purified GFP+ cells from uninjured skin (Figure S5C), these data suggest that committed adipocytes may not survive scRNA-seq processing and that a rare population (< 0.5% of mesenchymal cells, Figure S5A) of dermal cells are derived from adipocytes in telogenic skin (Zhang et al., 2019). However, given that Adipoq-traced myofibroblasts are absent from wound beds in mice that cannot undergo adipocyte lipolysis, our data strongly support that mature adipocytes undergo a fate switch to a myofibroblast identity, which is consistent with other reports of adipocyte plasticity (Marangoni et al., 2015; Zhang et al., 2019b). Taken together, these data suggest that following lipolysis, adipocytes lose their adipogenic identity and become myofibroblasts that support skin repair through the generation of ECM molecules (Fig. 7D).
DISCUSSION
Recent studies have identified critical functions of adipocytes to tissue function in the skin, mammary gland and bone marrow (Festa et al., 2011; Plikus et al., 2008; Shook et al., 2018; Zhang et al., 2015; Zhou et al., 2017a; Zwick et al., 2018). We have previously shown that inhibiting lineage commitment of adipocyte precursors, through PPARg inhibition, impairs wound bed fibroblast repopulation (Schmidt and Horsley, 2013). Here, we demonstrate that lipolysis of dermal adipocytes contributes to wound healing by regulating inflammatory macrophage infiltration. In addition to mobilizing lipid stores, the wound environment induces a change in adipocyte cellular plasticity that allows adipocyte-derived cells at the wound edge to become ECM-producing wound bed myofibroblasts during the proliferative phase of repair (Figure 7D). Our data resonate with other studies illustrating the ability of adipocyte-derived lipids to drive inflammatory states of adipose tissue (Alvarez-Curto and Milligan, 2016; Masoodi et al., 2015; Sohn et al., 2018) and promote tumorigenesis (Nieman et al., 2019; Zhang et al., 2018b).
Interestingly, activation of lipolysis through starvation or pharmacological adrenergic activation of adipose tissue is correlated with infiltration of macrophages into visceral adipose depots (Kosteli et al., 2010). Additionally, disrupting perilipin proteins that protect triglycerides from enzymatic breakdown leads to increased lipid mobilization and increased numbers of adipose tissue macrophages (Norman et al., 2018; Sohn et al., 2018; Zhou et al., 2017b). Furthermore, adipocyte deletion of Atgl results in loss of acute inflammation in visceral adipose tissue (Schoiswohl et al., 2015). Adipocyte-derived lipids might be directly signaling to macrophages, since macrophages express multiple fatty acid receptors and transporters (Alvarez-Curto and Milligan, 2016; Johnson et al., 2016), and inflammatory macrophages rely on glycolysis for energy (Biswas and Mantovani, 2012; Kominsky et al., 2010). Interestingly, heat-inactivated adipocyte-conditioned media increases monocyte/macrophage migration in vitro (Sohn et al., 2018), demonstrating that lipids can promote macrophage migration. In this study, we have observed that inhibiting dermal adipocyte lipolysis decreases the quantity of medium chain FAs that could promote macrophage migration through GPR84 signaling (Recio et al., 2018; Suzuki et al., 2013). Future studies elucidating the mechanisms by which FAs drive macrophage recruitment to tissues may lead to therapies for multiple inflammatory conditions.
Additionally, our work underscores the need to investigate dermal adipose tissue in human diabetic patients. Diabetic humans and mice have increased basal but impaired stimulated lipolysis in visceral and subcutaneous fat (Berndt et al., 2008; Bialesova et al., 2017; Girousse et al., 2013; Langin et al., 2005; Schoiswohl et al., 2015; Verboven et al., 2018). Similar alterations in lipolysis of diabetic dermal adipocytes would result in an inability to sense elevated FA levels at the site of injury and could contribute to impaired diabetic wound healing (Boniakowski et al., 2017; Doupis et al., 2010; Eming et al., 2014; Mirza et al., 2013; Seitz et al., 2010).
While tissue repair involves the plasticity of epithelial cell lineages (Burclaff and Mills, 2018; Donati et al., 2017; Ge et al., 2017; Ito et al., 2005; Sada et al., 2016; Tata et al., 2013), our data indicate that repair involves plasticity within mesenchymal lineages with adipocytes forming myofibroblasts. Adipocytes can undergo dramatic changes in size in the mammary gland during lactation and involution (Wang et al., 2018; Zwick et al., 2018), in visceral adipose tissue during fasting (Birsoy et al., 2008), and in the skin during the hair cycle (Festa et al., 2011; Zhang et al., 2019b), but they remain restricted to the white adipocyte lineage. Adipocytes grown in a floating “ceiling culture” have been reported to down-regulate adipocyte-specific genes, become fibroblast-like and enter the cell cycle, yet they retain the ability to become mature adipocytes (Kajita et al., 2013; Liao et al., 2015; Marangoni et al., 2015; Martins et al., 2015; Ono et al., 2011; Shen et al., 2011; Sugihara et al., 1986; Zhang et al., 2000). While mature adipocytes acquire a fibroblast-like transcriptional signature after high fat dieting (Jones et al., 2020), and adiponectin-expressing cells become myofibroblasts in mice in a bleomycin-induced skin fibrosis model (Marangoni et al., 2015; Zhang et al., 2019a), whether they derive from immature or mature adipocyte lineage cells is not clear. Our data indicate that adipocytes that undergo lipolysis can become myofibroblasts in skin wounds. It will be interesting to determine if a change in adipocyte cellular identity contributes to the loss of dermal adipose tissue with age (Rivera-Gonzalez et al., 2016; Tchkonia et al., 2010; Zhang et al., 2018a) and the long-term cellular identity and function of adipocyte-derived cells in tissue repair and fibrosis.
Together, our data highlight a role for adipocytes in tissue repair by promoting the inflammatory response and changing their cellular identity. It will be interesting to determine if age- and obesity-related changes in dermal adipocyte lipolysis and FAs contribute to reduced macrophage recruitment in aged skin, in response to S. aureus infection (Brubaker et al., 2013; Gosain and DiPietro, 2004; Pozzi et al., 2015; Zhang et al., 2018a) and non-healing wounds in diabetics (Krzyszczyk et al., 2018; Kuk et al., 2009; Tchkonia et al., 2010; Zhao et al., 2016). Future lines of investigation examining adipocyte lipolysis and adipocyte identity in conditions with skin inflammation and adipose tissue atrophy, such as tumorigenesis and fibrosis, could lead to breakthroughs in our treatment of human diseases.
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for reagents and resources should be directed to and will be fulfilled by the Lead Contact, Valerie Horsley (valerie.horsley@yale.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Wild type C57BL6/J mice were purchased from Charles River. B6;FVB-Tg(Adipoqcre)1Evdr/J (AdipoqCre); B6.129-Tg(Adipoq-cre/Esr1*)1Evdr/J (AdipoqCreER); 6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J (Rosa26CreER); B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (mT/mG); B6N.129S-Pnpla2tm1Eek/J (Atglfl/fl); and C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J (iDTR) mice were purchased from The Jackson Laboratories. Mice were maintained through routine breeding in an Association for Assessment and Accreditation of Laboratory Animal Care (AALAC)-accredited animal facility at Yale University. Both male and female mice were used in these studies, with no significant differences observed between the sexes. Animals were maintained on a standard chow diet ad libitum (Harlan Laboratories, 2018S) in 12-hour light/dark cycling. Two or three injured mice were housed per cage. All experimental procedures were approved and in accordance with the Institutional Animal Care and Use Committee.
Tissue Explants
Tissue explants were freshly dissected and immediately grown at 37°C with 5% CO2 in Lab-Tek Chamber Slides (Nunc).
METHOD DETAILS
Cell ablation, lipolysis inhibition and lineage tracing
To preferentially deplete dermal adipocytes, AdipoqCre; mT/mG; iDTR mice received an intradermal injection of 10μl of 20μg/μl diphtheria toxin (DT) (Sigma) in PBS. To preferentially inhibit lipolysis in dermal adipocytes, 100μl of 5mg/mL tamoxifen (Sigma Aldrich) was topically administered to shaved dorsal skin of AdipoqCreER; mT/mG; Atglfl/fl mice at indicated time points relative to wounding. For experiments using intraperitoneal (i.p.) tamoxifen administration, 100μl of 30mg/mL tamoxifen (Sigma Aldrich) in sesame oil was injected. All transgenic mice used for dermal adipocyte ablation and lipolysis inhibition contained the mT/mG allele to confirm Cre activity, and Cre-negative littermate controls were used for each experiment.
For EdU experiments, 50mg/kg of EdU (Invitrogen) was injected intraperitoneally at indicated time points and detected per manufacturer protocols. Detection of EdU-incorporating cells was performed using Click-it EdU Imaging or Flow Cytometry Assay kits (Invitrogen).
Dorsal skin excision
7–9-week-old mice were wounded during the telogen phase of hair cycling. Mice were anesthetized using isoflurane and four or six full-thickness wounds, at least 4mm apart, were made on shaved back skin using a 4mm biopsy punch (Accuderm). In experiments comparing different biopsy punch sizes, 2mm, 4mm, 6mm and 8mm biopsy punches (Accuderm) were used to make 2 full-thickness wounds on shaved back skin. Animals were sacrificed at noted intervals after injury and wound beds were processed for subsequent analysis.
Immunofluorescence and imaging
Inguinal subcutaneous white adipose tissue (SWAT) and perigonadal visceral white adipose tissue (GWAT) was dissected (Mann et al., 2014) and fixed in 4% paraformaldehyde for 2 hours at room temperature. Fixed adipose tissue was then washed with 1X PBS three times for 10 minutes each, incubated overnight at 4°C in 30% sucrose, then embedded in O.C.T. compound as previously described (Zwick et al., 2018). Mouse skin and wound beds were embedded in O.C.T. and wound beds were sectioned through their entirety to identify the center. 14μm cryosections were processed as previously described (Shook et al., 2016) and stained with antibodies listed in the Key Resources Table. LipidTOX (Invitrogen) was diluted in 1X PBS and staining was performed per the manufacturer’s protocol. To examine changes in adipocyte size during wound healing, tissue was fixed in 4% formaldehyde prior to embedment in O.C.T. as previously described (Salz and Driskell, 2017), then 40μm cryosections were cut and processed for immunofluorescent staining. Histological quantification for each wound bed was conducted on the three central-most sections and the averages from two wounds were averaged for each animal. Composite images were acquired using the tiles module on a Zeiss AxioImager M1 (Zeiss) equipped with an Orca camera (Hamamatsu). The percentage of the wound bed covered by ITGA6 staining (re-epithelialization), width of the wound bed, and ER-TR7 corrected total fluorescence were calculated from the 3 central most tissue sections using ImageJ software (National Institutes of Health, Bethesda, MD) as described previously (Schmidt and Horsley, 2013; Shook et al., 2018). Revascularization (CD31+) and fibroblast repopulation (SMA) were calculated using Adobe Photoshop to measure the total pixels positive for antibody staining divided by the total number of pixels in wound beds. Tissue resident macrophages were quantified by counting the number of CD68+ cells 850μm from the wound edge and between the bottom of the hair follicle (telogen) and the top of the panniculus carnosus.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC/eFluor 780 anti-mouse CD45 rat monoclonal (Clone 30-F11) | eBioscience | Cat# 47-0451-82 RRID: AB_1548781 |
| Alexa Fluor 700 anti-mouse CD11b rat monoclonal (Clone M1/70) | eBioscience | Cat# 56-0112-82 RRID: AB_657585 |
| eFluor 450 anti-mouse F4/80 rat monoclonal (Clone BM8) | eBioscience | Cat# 48-4801-82 RRID: AB_1548747 |
| PE/Cy7 anti-mouse Ly6G rat monoclonal (clone 1A8) | Biolegend | Cat# 127618 RRID: AB_1877261 |
| APC anti-mouse Ly6C rat monoclonal (clone HK1.4) | eBioscience | Cat# 17–5932 RRID: AB_1724155 |
| Alexa Fluor 488 anti-mouse CD206 rat monoclonal (clone C068C2) | Biolegend | Cat# 141710 RRID: AB_10900445 |
| eFluor 450 anti-mouse MHCII rat monoclonal (clone M5/114.15.2) | eBioscience | Cat# 48-5321-82 RRID: AB_1272204 |
| PE/Cy7 anti-mouse CD11c Armenian hamster monoclonal (clone N418) | Biolegend | Cat# 117318 RRID: AB_493568 |
| APC anti-mouse EpCam rat monoclonal (clone G8.8) | BD Biosciences | Cat# 563478 RRID: AB_2738234 |
| PerCp/Cy5.5 anti-mouse CD64 rat monoclonal (clone X54–5/7.1) | Biolegend | Cat# 139308 RRID: AB_2561963 |
| FITC anti-mouse CD3e Armenian hamster monoclonal (clone 145–2C11) | eBioscience | Cat# 11-0031-82 RRID: AB_464882 |
| PerCp anti-mouse CD4 rat monoclonal (clone GK1.5) | Biolegend | Cat# 100434 RRID: AB_893324 |
| APC anti-mouse CD8a rat monoclonal (clone 53–6.7) | eBioscience | Cat# 17-0081-83 RRID: AB_469336 |
| PE anti-mouse gd-TCR Armenian hamster monoclonal (clone GL3) | BD Biosciences | Cat# 553178 RRID: AB_394689 |
| Alexa Fluor 700 anti-mouse CD29 Armenian hamster monoclonal (clone HMbeta1–1) | Biolegend | Cat# 102218 RRID: AB_493711 |
| Brilliant Violet 421 anti-mouse CD34 rat monoclonal (clone MEC14.7) | Biolegend | Cat# 119321 RRID: AB_10900980 |
| Brilliant Violet 500 anti-mouse Ly-6A/E (SCA1) rat monoclonal (clone D7) | BD Biosciences | Cat# 561229 RRID: AB_10561841 |
| PE/Cy7 anti-mouse CD26 rat monoclonal (clone H194–112) | Biolegend | Cat# 137810 RRID: AB_2564312 |
| Anti-mouse CD45 Alexa Fluor 700 | eBioscience | Cat# 56-0451-82 RRID: AB_891454 |
| APC anti-mouse gdTCR Armenian hamster monoclonal (GL3) | Biolegend | Cat# 118116 RRID: AB_1731813 |
| PE anti-mouse CD3e Armenian hamster (145–2C11) | BD Biosciences | Cat# 553063 RRID: AB_394596 |
| APC-eFluor 780 anti-mouse CD3e Armenian hamster (145–2C11) | eBioscience | Cat# 47-0031-82 RRID: AB_11149861 |
| APC-Fire750 anti-mouse CD31 rat monoclonal (390) | Biolegend | Cat# 102434 RRID: AB_2629683 |
| Rat anti-mouse Foxp3 eFluor 450 | eBioscience | Cat# 48-5773-82 RRID: AB_1518812 |
| Brilliant Violet 650 anti-mouse CD4 | Biolegend | Cat# 100545 RRID: AB_11126142 |
| PE-Cyanine7 rat anti-mouse IL-17A | eBioscience | Cat# 25-7177-80 RRID: AB_10717952 |
| FITC rat anti-mouse IFN (XMG1.2) | eBioscience | Cat# 11-7311-82 RRID: AB_465412 |
| Brilliant Violet 605 anti-mouse CD8a | Biolegend | Cat# 100743 RRID: AB_2561352 |
| Anti-Mouse/Rat CD278 (ICOS) FITC | eBioscience | Cat# 11-9949-80 RRID: AB_465457 |
| PerCP-eFluor 710 anti-mouse CD27 Armenian hamster (LG.7F9) | eBioscience | Cat# 46-0271-82 RRID: AB_1834447 |
| APC anti-mouse CD103 Armenian hamster (2E7) | eBioscience | Cat# 17-1031-82 RRID: AB_1106992 |
| APC anti-rat CD140a (Pdgfra) (APA5) | Biolegend | Cat# 135908 RRID: AB_2043970 |
| Anti-Collagen I rabbit polyclonal | Abcam | Cat# ab34710 RRID: AB_731684 |
| Anti-Collagen III rabbit polyclonal | Abcam | Cat# ab7778 RRID: AB_306066 |
| Anti-Fibrillin 1 rabbit polyclonal | Novus | Cat# NBP1–84723 RRID: AB_11005848 |
| Anti-Fibronectin rabbit polyclonal | Abcam | Cat# ab23750 RRID: AB_447655 |
| Anti-alpha smooth muscle actin rabbit polyclonal | Abcam | Cat# ab5694 RRID: AB_2223021 |
| Anti-Perilipin1 goat polyclonal | Abcam | Cat# ab61682 RRID: AB_944751 |
| Anti-Pparg rabbit monoclonal (clone K.242.9) | Thermo | Cat# MA5–14889 RRID: AB_10985650 |
| Anti-Vimentin rabbit monoclonal (clone D21H3) | Cell Signaling | Cat# 5741 RRID: AB_10695459 |
| Anti-CD31 rat monoclonal (clone MEC13.3) | BD Bioscience | Cat# 550274 RRID: AB_393571 |
| Anti-mouse ITGA6 rat monoclonal (clone GoH3) | R&D Systems | Cat# MAB13501 RRID: AB_2128311 |
| Anti-ER-TR7 rat monoclonal | Abcam | Cat# ab51824 RRID: AB_881651 |
| Anti-GFP chicken polyclonal | Abcam | Cat# ab13970 RRID: AB_300798 |
| Anti-CD68 rabbit polyclonal | Abcam | Cat# ab125212 RRID: AB_10975465 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Diphtheria Toxin | Sigma | D0564 |
| Tamoxifen | Sigma | T5648 |
| LipidTOX | Invitrogen | H34477 |
| Liberase TM | Roche | 05401127001 |
| Sytox Orange | Invitrogen | S34861 |
| Sytox Blue | Invitrogen | S34857 |
| Sybr Green | Roche | 04887352001 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit | Invitrogen | C10419 |
| Click-iT EdU Alexa Fluor 647 Imaging Kit | Invitrogen | C10340 |
| Ovation RNA-seq System V2 | NuGEN | 7102 |
| Ovation Ultralow DR Multiplex System 1–8 | NuGEN | 0330 |
| Absolutely RNA Nanoprep Kit | Agilent | 400753 |
| RNeasy Plus Micro Kit | Qiagen | 74034 |
| Foxp3/Transcription Factor Staining Buffer Set | eBioscience | 00552300 |
| Cell Stimulation Cocktail | Tonbo Biosciences | TNF-4975 |
| Ghost Dye Violet 510 Live/Dead Stain | Tonbo Bioscience | Cat# 13-0870-T100 |
| Chromium i7 Multiplex Kit | 10x Genomics | PN-120262 |
| Chromium Single Cell B Chip Kit | 10x Genomics | PN-1000073 |
| Chromium Single Cell 3’ GEM Library and Gel Bead Kit v3 | 10x Genomics | PN-1000075 |
| Deposited Data | ||
| Bulk RNA-seq data | This study | GSE126514 |
| Single cell RNA-seq data | This study | GSE140512 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6;FVB-Tg(Adipoq-cre)1Evdr/J | The Jackson Laboratory | 010803 |
| Mouse: B6.129-Tg(Adipoq-cre/Esr1*)1Evdr/J | The Jackson Laboratory | 024671 |
| Mouse: 6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J | The Jackson Laboratory | 008463 |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | 007676 |
| Mouse: B6N.129S-Pnpla2tm1Eek/J | The Jackson Laboratory | 024278 |
| Mouse: C57BL/6-Gt(ROSA)26Sortm1(HBEGF)Awai/J | The Jackson Laboratory | 007900 |
| Mouse: C57BL/6 | Charles River | 027 |
| Oligonucleotides | ||
| PCR primers, see Table S3 | This study | N/A |
| Software and Algorithms | ||
| Fiji (ImageJ) | NIH | https://fiji.sc |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| FlowJo | FlowJo, LLC | https://www.flowjo.com |
| Ingenuity Pathway Analysis | Qiagen | https://www.qiagenbioinformatics.com |
| MATLAB | MathWorks | https://www.mathworks.com |
| GraphPad Prism | GraphPad Software, Inc | https://www.graphpad.com |
| HISAT2 2.1.0 | Kim et al., 2015 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| StringTie v1.3.3b | Pertea et al., 2015 | https://ccb.jhu.edu/software/stringtie/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Kallisto | Bray et al., 2016 | https://pachterlab.github.io/kallisto/ |
| Bustools | Melsted et al., 2019 | https://www.kallistobus.tools |
| Seurat 3.0 | Stuart et al., 2019 | https://satijalab.org/seurat/ |
Quantification of spatial distribution of cells in wound beds
In order to quantify the distribution of immunoreactivity relative to the wound edge, we created a custom MATLAB program. First, the edge and midline of the wound bed was manually labeled. Then, for each image, we set a threshold of 10% of the maximum intensity in that image. Pixels below this threshold were set to zero. Next, for every possible distance from the wound bed edge, the program calculated the mean intensity of the desired channel of every pixel at that distance. The outer and superficial wound edges were defined as the outermost points of the wound bed along the respective axis. The program used normalized distance units to account for different wound bed shapes. For every depth of the wound bed, the distance units were normalized according to the width of the wound bed at that depth, and for every point along the edge-midline axis, the distance was normalized according to the depth of the wound bed at that point.
Transmission electron microscopy (TEM)
TEM was performed in the Yale School of Medicine Center for Cellular and Molecular Imaging Electron Microscopy core facility. AdipoqCreER+; Atglfl/fl and AdipoqCreER−; Atglfl/fl mice were treated with tamoxifen as described above. Two days after tamoxifen administration, mice were wounded and then sacrificed 24 hours after injury by CO2 and immediately transcardially perfused using PBS, then 20–30ml of cold 4% PFA according to standard protocols (Xu et al., 2013; Zwick et al., 2018). The wound edge was dissected and cut into small pieces, approximately 1mm3 in volume. Tissue pieces were fixed in 2.5% glutaraldehyde/2% PFA in 0.1M sodium cacodylate buffer, pH 7.4, for 30 min at RT and 1.5 hours at 4 °C, and then rinsed in sodium cacodylate buffer three times. Samples were postfixed in 1% osmium tetroxide for 1 hour, rinsed and en bloc stained in aqueous 2% uranyl acetate for 1 hour followed by rinsing, dehydrating in an ethanol series to 100%, rinsing in 100% propylene oxide, infiltrating with Embed 812 resin (Electron Microscopy Sciences) and baking overnight at 60°C. Hardened blocks were cut using an ultramicrotome (UltraCut UC7; Leica). Ultrathin 60nm sections were collected and stained using 2% uranyl acetate and lead citrate for transmission microscopy. Carbon-coated grids were viewed on a transmission electron microscope (Tecnai BioTWIN; FEI) at 80kV. Images were taken using a CCD camera (Morada; Olympus) and iTEM (Olympus) software.
Flow cytometry and sorting
Mouse back skin and wound beds were dissected and digested into single cells using Liberase TM (Roche) and cells were suspended in FACS staining buffer (1% BSA in PBS with 2mM EDTA). To isolate mature adipocytes, digested samples were centrifuged at 300g for 4 minutes and floating adipocytes were isolated using a pipette (Church et al., 2014). For samples that required immunostaining, digested tissue was filter with a 70μm and 40μm cell strainer prior to centrifugation. Cell suspensions were stained with antibodies purchased from eBioscience, Biolegend and BD Bioscience in the Key Resources Table for 20–30 minutes on ice. Wound macrophages were defined as CD45+; CD11b+; F4/80+ cells and neutrophils as CD45+; CD11b+; F4/80−; Ly6G+ cells. Analysis of proliferation using EdU incorporation was performed using the Click-iT EdU Flow Cytometry Assay Kit per the manufacturer’s instructions (Invitrogen). To exclude dead cells, Sytox Orange or Sytox Blue (Invitrogen, 1:1000) was added immediately before analysis or sorting using a FACS Aria III with FACS DiVA software (BD Biosciences). Cells were sorted into 10% fetal bovine serum (FBS) in DMEM and flow cytometry analysis was performed using FlowJo Software (FlowJo).
To analyze T cell activation and cytokines using intracellular flow cytometry, skin was harvested, lightly defatted, minced with scissors and resuspended in a digestion mix with collagenase XI (2mg/ml; Sigma-Aldrich, catalog no. C9407), hyaluronidase (0.5mg/ml; Sigma-Aldrich, catalog no. H3506) and deoxyribonuclease (0.1mg/ml; Sigma-Aldrich, catalog no. DN25) in RPMI 1640 with 2% fetal calf serum and 1% penicillin-streptomycin. Skin was digested for 45 minutes in a 37°C shaking incubator at 225rpm, then washed and vortexed for 15 seconds before filtering through a 100μm strainer. Cell counts were performed using a Nucleocounter NC-200 (ChemoMetec). Cells were stimulated ex vivo with Cell Stimulation Cocktail (Tonbo Biosciences, catalog no. TNB-4975) for 4 hours before staining for flow cytometry analysis. Cells were stained for surface antigens and a live/dead marker (Ghost Dye Violet 510, Tonbo Biosciences) in FACS buffer (PBS with 2% fetal calf serum and 1% penicillin-streptomycin) for 30 minutes at 4°C. To stain for intracellular markers, cells were fixed and permeabilized using the Foxp3-Transcription Factor Staining Buffer set (eBioscience). Samples were run on a Fortessa (BD Biosciences) in the UCSF Flow Cytometry Core. FlowJo software (FlowJo LLC) was used to analyze flow cytometry data.
Lipid profiling
For quantitative lipid mass spectrometry of adipocyte lipid stores, skin or subcutaneous adipose tissue was digested with Liberase TM mixture as described above for flow cytometry. Released SVF cells were centrifuged and the floating adipocyte fraction was isolated, washed with PBS and snap frozen in liquid nitrogen for mass spectrometry. To analyze lipids during wound healing, 2mm of tissue around the wound edge was dissected with scissors and snap frozen. Total lipids were extracted and FA profiles quantified by GC/MS as previously described (Fraher et al., 2016; Rudolph et al., 2017). For eicosanoid quantification, snap frozen adipose tissue was pulverized and approximately 10–15mg was added to 2mL of 50% methanol containing internal heavy atom standards ([2H4]LTB4, [2H5]LTD4, [2H8]5–HETE, [2H4]PGF2α, [2H4]PGE2, [2H4]TXB2 (2ng each), [2H5]LTC4, [2H5]LTE4 and [2H4]6-keto-PGF1α (5ng each) and [2H8]AA, (10ng)), as previously described (Zarini et al., 2014). Samples were mechanically homogenized at a low setting, the lysate centrifuged at 4,000g for 15 minutes at 4°C and the supernatant was added to 6 mL of ultrapure water (final methanol = 12.5%). Strata-X 33u polymeric reverse-phase cartridges (60mg/mL, Phenomenex, Torrance, CA) were conditioned with 4mL of 100% methanol and equilibrated with 4mL of ultrapure water, after which the samples were allowed to pass through by gravity. Columns containing eicosanoids were washed with 4mL and then 1mL of ultrapure water, eicosanoids were eluted with 1mL of 100% methanol and samples were taken to dryness under N2 gas at room temperature. Lipid pellets were resuspended in 60μL of 65:35 acetonitrile/methanol vol/vol (solvent B) to which 40μL of 8.3mM acetic acid in ultrapure water brought to pH 5.7 with ammonium hydroxide (solvent A) was added and transferred to autosampler vials. An aliquot of each sample (20μL) was injected into a HPLC column (Accucore C18 50 × 3mm, 2.6μm, Thermo Scientific, Waltham, MA) and eluted at a flow rate of 300L/minute with a linear gradient of HPLC solvent B, which was increased from 45% to 75% in 6.5 minutes, to 98% in 1 minutes and held at 98% for a further 6.5 minutes before re-equilibration at 45% for 10 minutes. The HPLC system was directly interfaced into the electrospray source of a triple quadrupole mass spectrometer (API 3000, PE–Sciex) where mass spectrometric analysis was performed in negative-ion mode using MRM of specific m/z transitions and eicosanoids were quantified, as previously described (Zarini et al., 2014).
Wound bed explant culture
Wound beds were dissected from back skin and the hair follicle containing edge was removed. Bisected wound beds were cultured in 10% FBS in DMEM (ATCC) in chamber slides (ThermoFisher) for 2 days. Cells that migrated away from the center of the wound bed were immunostained as described above with GFP and vimentin antibodies.
RNA-seq and Quantitative Real-Time PCR
FACS-purified samples were digested using TRIzol LS (Invitrogen), RNA was extracted from the aqueous phase using the RNeasy Plus Micro Kit (Qiagen). cDNA was generated using equal amounts of total RNA with the Superscript III First Strand Synthesis Kit (Invitrogen) per manufacturer instructions. All quantitative real-time PCR was performed using SYBR green on a LightCycler 480 (Roche). Primers for specific genes are listed in Supplementary Table 3. Results were normalized to β-actin as previously described.
For bulk RNA-seq experiments, total RNA was obtained from low input FACS-isolated cellular subsets or floating adipocytes using the Absolutely RNA Nanoprep Kit (Agilent). Quality RNA samples were selected (RIN>7; Agilent 2100 Bioanalyzer) and RNA-seq was performed. From each cellular subset, 500pg-1ng of total RNA was purified and converted to cDNA using the Ovation RNA-seq System V2 and Ovation Ultralow DR Multiplex System (NuGEN) per manufacturer protocols. Samples were sequenced at the Yale Center for Genomic Analysis using 75bp single-end sequencing on an Illumina HiSeq 2500 according to Illumina protocols generating an average of 32 million paired-end reads per library.
Construction of 10X Genomic Single Cell 3’ RNA-Seq libraries and sequencing
First, single cell suspension in RT Master Mix was loaded on the Single Cell a Chip and partition with a pool of about 750,000 barcoded gel beads to form nanoliter-scale Gel Beads-In-Emulsions (GEMs). Upon dissolution of the Gel Beads in a GEM, the primers with the unique cell barcodes are released and mixed with cell lysate and Master Mix. Silane magnetic beads were used to remove leftover biochemical reagents and primers from the post GEM reaction mixture. Full-length, barcoded cDNA was amplified by PCR to generate sufficient mass for library construction using a Chromium Controller (10× Genomics). Enzymatic Fragmentation and Size Selection were used to optimize the cDNA amplicon size prior to library construction, which includes end Repair, A-tailing, adaptor Ligation, and sample indexing PCR to produces Illumina-ready sequencing libraries. The final libraries contain the P5 and P7 primers used in Illumina bridge amplification. A16bp 10× Barcode and 10bp UMI are encoded in Read 1, while Read 2 is used to sequence the cDNA fragment. For cell preparation and sequencing, sample concentrations are normalized to 2nM and loaded onto Illumina NovaSeq 6000 flow cells at a concentration that yields at least 2 billion of passing filter data per lane. Loading concentration for 10× libraries have been optimized to maximize both well occupancy and unique read output while limiting duplicates associated with patterned flow cell technology. Samples are sequenced according to Illumina and 10× Genomics protocols on an Illumina HiSeq 4000. Data generated during sequencing runs are simultaneously transferred to the YCGA high-performance computing cluster. A positive control (prepared bacteriophage Phi X library) provided by Illumina is spiked into every lane at a concentration of 1% to monitor sequencing quality in real time.
QUANTIFICATION AND STATISTICAL ANALAYSIS
RNA-sequencing Analysis
For bulk RNA-seq, each sequencing read, the first 6 and the last nucleotides were trimmed to the point where the Phred score of an examined base fell below 20 using in-house scripts. If, after trimming, the read was shorter than 45bp, the whole read was discarded. Trimmed reads were mapped to the mouse reference genome (mm10) with HISAT2 v2.1.0 (Kim et al., 2015) reporting alignments tailored for transcript assemblers. Alignments with quality score below 20 were excluded from further analysis. Gene counts were produced with StringTie v1.3.3b (Pertea et al., 2015) and the Python script “prepDE.py” provided in the package. StringTie was limited to reads matching the reference annotation GENCODE v27(Harrow et al., 2012). After obtaining the matrix of read counts, differential expression analysis was conducted and normalized counts were produced using DESeq2 (Love et al., 2014). P-values were adjusted for multiple testing using the Benjamini-Hochberg procedure (Benjamini et al., 2001). Significantly differentially expressed genes (log2 fold change ≥ 1, FDR-adjusted p<0.05) were analyzed using Ingenuity Pathway Analysis software suite (Qiagen).
For single cell RNA-seq, 8,234 cells were sequenced from uninjured skin and >10,000 cells were sequenced from each wound bed (16,332 cells from a male sample and 10,999 from a female sample). For all samples, the mean reads per cell of >45,000 with >3000 genes per cell. Reads were demultiplexed and gene-level quantification were quantified using the Kallisto and Bustools suite (Bray et al., 2016; Melsted et al., 2019) with a custom annotation combining the GENCODE vM22 mouse gene set with eGFP and tdTomato sequences present in the mT/mG mouse. Processing of scRNA-Seq data was conducted using version 3 of the Seurat package (Butler et al., 2018; Stuart et al., 2019). Cells were filtered for detection of at least 1000 genes and at least 3000 total counts per cell (Fig S7A–C), gene expression counts were normalized using the sctransform approach (Hafemeister and Satija, 2019), and batch effects were removed using the anchor-based Canonical Correlation Analysis approach (Stuart et al., 2019). Clustering was performed using the Louvain algorithm and marker gene detection was performed with the Wilcoxon rank sum test, both using the Seurat package. To identify GFP positive cells, we required expression of eGFP at 700 UMI counts per million–a level similar to the expression of the eGFP in bulk RNA-Seq from the sorted GFP+ population–and at least 10 reads mapping to eGFP and no detectable tomato reads (Figure S7D). For all cells meeting these requirements, at least 95% of reads mapping to eGFP or tdTomato mapped to eGFP (Figure S7D).
Statistics
To determine significance between two groups, comparisons were made using Student’s t-test. Analyses across multiple groups were made using a one-way ANOVA with Bonferroni’s post hoc using GraphPad Prism for Mac (GraphPad Software, La Jolla, CA) with significance set at p < 0.05.
DATA AND SOFTWARE AVAILABILITY
Sequencing and analysis data were deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE126514 and GSE140512.
Supplementary Material
Table S1. Ingenuity Pathway Analysis of differentially expressed molecules in fibroblast subsets. Related to Figure 7 and Figure S7.
Table S2. Ingenuity Pathway Analysis associated diseases and functions of fibroblast subsets. Related to Figure 7 and Figure S7.
Table S3. Mouse qPCR primers. Related to STAR Methods.
Highlights:
Inhibiting dermal adipocyte lipolysis reduces inflammatory wound bed macrophages
Wound edge adipocytes dedifferentiate within hours after injury
Adipocyte lipolysis is needed for dedifferentiated adipocytes to populate wound beds
Dedifferentiated adipocytes generate wound bed myofibroblasts after injury
ACKNOWLEDGEMENTS
This work was supported in part by NIH grants to V.H. from NIAMS (AR060295 & AR069550) and NIA through the pilot project grants from the Claude D. Pepper Older Americans Independence Center at Yale (NIA P30AG21342) awarded to V.H. MCR is supported by NIDDK (K01DK109079). This research was supported by the New York Stem Cell Foundation. RNA-seq data are available at the Gene Expression Omnibus (GEO) at www.ncbi.nlm.nih.gov/geo, under accession numbers GSE126514 and GSE140512. We thank members of the Horsley lab for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors have no competing interests.
REFERENCES
- Alvarez-Curto E, and Milligan G (2016). Metabolism meets immunity: The role of free fatty acid receptors in the immune system. Biochemical Pharmacology 114, 3–13. [DOI] [PubMed] [Google Scholar]
- Bani D, Li AQ, Freschi G, and Russo GL (2013). Histological and Ultrastructural Effects of Ultrasound-induced Cavitation on Human Skin Adipose Tissue. Plastic and Reconstructive Surgery Global Open 1, e41–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos S, Stojadinovic O, Golinko MS, Brem H, and Tomic-Canic M (2008). Growth factors and cytokines in wound healing. Wound Repair and Regeneration 16, 585–601. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, and Golani I (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res 125, 279–284. [DOI] [PubMed] [Google Scholar]
- Berndt J, Kralisch S, Klöting N, Ruschke K, Kern M, Fasshauer M, Schön M, Stumvoll M, and Blüher M (2008). Adipose Triglyceride Lipase Gene Expression in Human Visceral Obesity. Exp Clin Endocrinol Diabetes 116, 203–210. [DOI] [PubMed] [Google Scholar]
- Bialesova L, x000E9 AXK, Petrus P, Sinha I, Laurencikiene J, Zhao C, Wright KD, Arner P, and Dahlman I (2017). Epigenetic Regulation of PLIN1 in Obese Women and its Relation to Lipolysis. Sci. Rep 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Soukas A, Torrens J, Ceccarini G, Montez J, Maffei M, Cohen P, Fayzikhodjaeva G, Viale A, Socci ND, et al. (2008). Cellular program controlling the recovery of adipose tissue mass: An in vivo imaging approach. Proc Natl Acad Sci USA 105, 12985–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, and Mantovani A (2012). Orchestration of Metabolism by Macrophages. Cell Metabolism 15, 432–437. [DOI] [PubMed] [Google Scholar]
- Boniakowski AE, Kimball AS, Jacobs BN, Kunkel SL, and Gallagher KA (2017). Macrophage-Mediated Inflammation in Normal and Diabetic Wound Healing. The Journal of Immunology 199, 17–24. [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Brubaker AL, Rendon JL, Ramirez L, Choudhry MA, and Kovacs EJ (2013). Reduced neutrophil chemotaxis and infiltration contributes to delayed resolution of cutaneous wound infection with advanced age. The Journal of Immunology 190, 1746–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burclaff J, and Mills JC (2018). Plasticity of differentiated cells in wound repair and tumorigenesis, part II: skin and intestine. Disease Models & Mechanisms 11, dmm035071–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church CD, Berry R, and Rodeheffer MS (2014). Isolation and study of adipocyte precursors. Meth. Enzymol 537, 31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, and Forster I (1999). Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research 8, 265–277. [DOI] [PubMed] [Google Scholar]
- Crane MJ, Daley JM, van Houtte O, Brancato SK, Henry WL, and Albina JE (2014). The Monocyte to Macrophage Transition in the Murine Sterile Wound. PLoS ONE 9, e86660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhall S, Wijesinghe DS, Karim ZA, Castro A, Vemana HP, Khasawneh FT, Chalfant CE, and Martins-Green M (2015). Arachidonic acid-derived signaling lipids and functions in impaired healing. Wound Repair and Regeneration 23, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, Kayikci M, Russell R, Kretzschmar K, Mulder KW, et al. (2017). Wounding induces dedifferentiation of epidermal Gata6+ cells and acquisition of stem cell properties. Nat Cell Biol 19, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupis J, Rahangdale S, Gnardellis C, Pena SE, Malhotra A, and Veves A (2010). Effects of Diabetes and Obesity on Vascular Reactivity, Inflammatory Cytokines, and Growth Factors. Obesity 19, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Jahoda CAB, Chuong C-M, Watt FM, and Horsley V (2014). Defining dermal adipose tissue. Exp. Dermatol 23, 629–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferrón SR, Herault Y, Pavlovic G, Ferguson-Smith AC, et al. (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann TO, Kumari M, Haas JT, Farese RV, Zimmermann R, Lass A, and Zechner R (2012). Studies on the substrate and stereo/regioselectivity of adipose triglyceride lipase, hormone-sensitive lipase, and diacylglycerol-O-acyltransferases. Journal of Biological Chemistry 287, 41446–41457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Martin P, and Tomic-Canic M (2014). Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6, 265sr6–265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR (2006). Transcriptional control of adipocyte formation. Cell Metabolism 4, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante CJ, Pinhal-Enfield G, Elson G, Cronstein BN, Hasko G, Outram S, and Leibovich SJ (2013). The Adenosine-Dependent Angiogenic Switch of Macrophages to an M2-Like Phenotype is Independent of Interleukin-4 Receptor Alpha (IL-4Rα) Signaling. Inflammation 36, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa E, Fretz J, Berry R, Schmidt B, Rodeheffer M, Horowitz M, and Horsley V (2011). Adipocyte Lineage Cells Contribute to the Skin Stem Cell Niche to Drive Hair Cycling. Cell 146, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraher D, Sanigorski A, Mellett NA, Meikle PJ, Sinclair AJ, and Gibert Y (2016). Zebrafish Embryonic Lipidomic Analysis Reveals that the Yolk Cell Is Metabolically Active in Processing Lipid. CellReports 14, 1317–1329. [DOI] [PubMed] [Google Scholar]
- Franz A, Wood W, and Martin P (2018). Fat Body Cells Are Motile and Actively Migrate to Wounds to Drive Repair and Prevent Infection. Dev Cell 44, 460–470.e463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frühbeck G, Méndez-Giménez L, Fernández-Formoso J-A, Fernández S, and Rodríguez A (2014). Regulation of adipocyte lipolysis. Nutr. Res. Rev 27, 63–93. [DOI] [PubMed] [Google Scholar]
- Futagami A, Ishizaki M, Fukuda Y, Kawana S, and Yamanaka N (2002). Wound Healing Involves Induction of Cyclooxygenase-2 Expression in Rat Skin. Lab. Invest 82, 1503–1513. [DOI] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CP-J, Polak L, Yuan S, Elemento O, et al. (2017). Stem Cell Lineage Infidelity Drives Wound Repair and Cancer. Cell 169, 636–642.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girousse A, Tavernier G, Valle C, Moro C, Mejhert N, Dinel A-L, Houssier M, Roussel B, Besse-Patin A, Combes M, et al. (2013). Partial Inhibition of Adipose Tissue Lipolysis Improves Glucose Metabolism and Insulin Sensitivity Without Alteration of Fat Mass. PLoS Biol 11, e1001485–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren I, Allmann N, Yogev N, Schürmann C, Linke A, Holdener M, Waisman A, Pfeilschifter J, and Frank S (2010). A Transgenic Mouse Model of Inducible Macrophage Depletion. Ajpa 175, 132–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosain A, and DiPietro LA (2004). Aging and wound healing. World J Surg 28, 321–326. [DOI] [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nature Communications 10, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C, and Satija R (2019). Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. bioRxiv 3, 861–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. (2012). GENCODE: The reference human genome annotation for The ENCODE Project. Genome Research 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himms-Hagen J, Melnyk A, Zingaretti MC, Ceresi E, Barbatelli G, and Cinti S (2000). Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. AJP: Cell Physiology 279, C670–C681. [DOI] [PubMed] [Google Scholar]
- Itabe H, Yamaguchi T, Nimura S, and Sasabe N (2017). Perilipins: a diversity of intracellular lipid droplet proteins. 1–11. [DOI] [PMC free article] [PubMed]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, and Cotsarelis G (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11, 1351–1354. [DOI] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, and Cotsarelis G (2007). Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320. [DOI] [PubMed] [Google Scholar]
- Johnson AR, Qin Y, Cozzo AJ, Freemerman AJ, Huang MJ, Zhao L, Sampey BP, Milner JJ, Beck MA, Damania B, et al. (2016). Metabolic reprogramming through fatty acid transport protein 1 (FATP1) regulates macrophage inflammatory potential and adipose inflammation. Molecular Metabolism 5, 506–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JEC, Rabhi N, Orofino J, Gamini R, Perissi V, Vernochet C, and Farmer SR (2020). The Adipocyte Acquires a Fibroblast-Like Transcriptional Signature in Response to a High Fat Diet. Sci. Rep 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajita K, Mori I, Kitada Y, Taguchi K, Kajita T, Hanamoto T, Ikeda T, Fujioka K, Yamauchi M, Okada H, et al. (2013). Small proliferative adipocytes: identification of proliferative cells expressing adipocyte markers. Endocr. J 60, 931–939. [DOI] [PubMed] [Google Scholar]
- Kasza I, Suh Y, Wollny D, Clark RJ, Roopra A, Colman RJ, MacDougald OA, Shedd TA, Nelson DW, Yen M-I, et al. (2014). Syndecan-1 Is Required to Maintain Intradermal Fat and Prevent Cold Stress. PLoS Genet 10, e1004514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall AC, and Nicolaou A (2013). Progress in Lipid Research. Progress in Lipid Research 52, 141–164. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nature Publishing Group 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominsky DJ, Campbell EL, and Colgan SP (2010). Metabolic Shifts in Immunity and Inflammation. Journal of Immunology (Baltimore, Md. : 1950) 184, 4062–4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosteli A, Sugaru E, Haemmerle G, Martin JF, Lei J, Zechner R, and Ferrante AW Jr. (2010). Weight loss and lipolysis promote a dynamic immune response in murine adipose tissue. Journal of Clinical Investigation 120, 3466–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyszczyk P, Schloss R, Palmer A, and Berthiaume F (2018). The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front. Physiol 9, 165–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk JL, Saunders TJ, Davidson LE, and Ross R (2009). Age-related changes in total and regional fat distribution. Ageing Research Reviews 8, 339–348. [DOI] [PubMed] [Google Scholar]
- Langin D, Dicker A, Tavernier G, Hoffstedt J, Mairal A, Rydén M, Arner E, Sicard A, Jenkins CM, Viguerie N, et al. (2005). Adipocyte lipases and defect of lipolysis in human obesity. Diabetes 54, 3190–3197. [DOI] [PubMed] [Google Scholar]
- Leibovich SJ, and Ross R (1975). The role of the macrophage in wound repair. A study with hydrocortisone and antimacrophage serum. Ajpa 78, 71–100. [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zeng Z, Lu F, Dong Z, Chang Q, and Gao J (2015). In Vivo Dedifferentiation of Adult Adipose Cells. PLoS ONE 10, e0125254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 31–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Muller W, Roers A, and Eming SA (2010). Differential Roles of Macrophages in Diverse Phases of Skin Repair. Journal of Immunology (Baltimore, Md. : 1950) 184, 3964–3977. [DOI] [PubMed] [Google Scholar]
- Macedo L, Pinhal-Enfield G, Alshits V, Elson G, Cronstein BN, and Leibovich SJ (2010). Wound Healing Is Impaired in MyD88-Deficient Mice. Ajpa 171, 1774–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann A, Thompson A, Robbins N, and Blomkalns AL (2014). Localization, Identification, and Excision of Murine Adipose Depots. JoVE 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni RG, Korman BD, Wei J, Wood TA, Graham LV, Whitfield ML, Scherer PE, Tourtellotte WG, and Varga J (2015). Myofibroblasts in murine cutaneous fibrosis originate from adiponectin-positive intradermal progenitors. Arthritis Rheumatol 67, 1062–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins V, De Los Santos FG, Wu Z, Capelozzi V, Phan SH, and Liu T (2015). FIZZ1-Induced Myofibroblast Transdifferentiation from Adipocytes and Its Potential Role in Dermal Fibrosis and Lipoatrophy. Ajpa 185, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoodi M, Kuda O, Rossmeisl M, Flachs P, and Kopecky J (2015). Lipid signaling in adipose tissue: Connecting inflammation & metabolism. BBA - Molecular and Cell Biology of Lipids 1851, 503–518. [DOI] [PubMed] [Google Scholar]
- Melsted P, Booeshaghi AS, Gao F, Beltrame E, Lu L, Hjorleifsson KE, Gehring J, and Pachter L (2019). Modular and efficient pre-processing of single-cell RNA-seq. bioRxiv 20, 65–16. [Google Scholar]
- Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364, eaav2501–eaav2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza RE, Fang MM, Ennis WJ, and Koh TJ (2013). Blocking interleukin-1β induces a healing-associated wound macrophage phenotype and improves healing in type 2 diabetes. Diabetes 62, 2579–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza R, DiPietro LA, and Koh TJ (2010). Selective and Specific Macrophage Ablation Is Detrimental to Wound Healing in Mice. Ajpa 175, 2454–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Perfield JW II, Obin MS, and Greenberg AS (2008). Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. J. Cell. Biochem 105, 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra J, Mason MM, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, et al. (1998). Life without white fat: a transgenic mouse. Genes Dev 12, 3168–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AM, Loy DE, Lawson JA, Katseff AS, FitzGerald GA, and Garza LA (2013). Prostaglandin D2 Inhibits Wound-Induced Hair Follicle Neogenesis through the Receptor, Gpr44. J. Invest. Dermatol 133, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou A (2013). Prostaglandins, Leukotrienes and Essential Fatty Acids. Prostaglandins, Leukotrienes and Essential Fatty Acids 88, 131–138. [DOI] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GOKS, et al. (2019). Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JE, Aung HH, Wilson DW, and Rutledge JC (2018). Inhibition of perilipin 2 expression reduces pro-inflammatory gene expression and increases lipid droplet size. Food Funct. 13, 544–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H, Oki Y, Bono H, and Kano K (2011). Biochemical and Biophysical Research Communications. Biochemical and Biophysical Research Communications 407, 562–567. [DOI] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, His T-C, et al. (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science aai8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, la Cruz de D, Baker RE, Maini PK, Maxson R, and Chuong C-M (2008). Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzi C, Lofano G, Mancini F, Soldaini E, Speziale P, De Gregorio E, Rappuoli R, Bertholet S, Grandi G, and Bagnoli F (2015). Phagocyte subsets and lymphocyte clonal deletion behind ineffective immune response to Staphylococcus aureus. FEMS Microbiol. Rev 39, 750–763. [DOI] [PubMed] [Google Scholar]
- Ramachandran P, Pellicoro A, Vernon MA, Boulter L, Aucott RL, Ali A, Hartland SN, Snowdon VK, Cappon A, Gordon-Walker TT, et al. (2012). Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA 109, E3186–E3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recio C, Lucy D, Purvis GSD, Iveson P, Zeboudj L, Iqbal AJ, Lin D, O’Callaghan C, Davison L, Griesbach E, et al. (2018). Activation of the Immune-Metabolic Receptor GPR84 Enhances Inflammation and Phagocytosis in Macrophages. Front. Immunol 9, 81–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman ML, and Gavrilova O (2000). A-ZIP/F-1 mice lacking white fat: a model for understanding lipoatrophic diabetes. Int J Obes Relat Metab Disord 24 Suppl 4, S11–S14. [DOI] [PubMed] [Google Scholar]
- Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, et al. (2015). Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348, aaa2151–aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Gonzalez GC, Shook BA, and Horsley V (2017). PDGFA regulation of dermal adipocyte stem cells. Stem Cell Investig. 4, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Gonzalez GC, Shook BA, Andrae J, Holtrup B, Bollag K, Betsholtz C, Rodeheffer MS, and Horsley V (2016). Skin Adipocyte Stem Cell Self-Renewal Is Regulated by a PDGFA/AKT-Signaling Axis. Stem Cell 19, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford JJ (2014). Mouse Models of Lipodystrophy and Their Significance in Understanding Fat Regulation (Elsevier Inc.). [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, and Friedman JM (2008). Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249. [DOI] [PubMed] [Google Scholar]
- Rodero MP, Hodgson SS, Hollier B, Combadiere C, and Khosrotehrani K (2012). Reduced Il17a Expression Distinguishes a Ly6c. Journal of Investigative Dermatology 133, 783–792. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2000). Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol 16, 145–171. [DOI] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2006). Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MC, Young BE, Lemas DJ, Palmer CE, Hernandez TL, Barbour LA, Friedman JE, Krebs NF, and MacLean PS (2017). Early infant adipose deposition is positively associated with the n-6 to n-3 fatty acid ratio in human milk independent of maternal BMI. Int J Obes Relat Metab Disord 41, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Jacob F, Leung E, Wang S, White BS, Shalloway D, and Tumbar T (2016). Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat Cell Biol 18, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz L, and Driskell RR (2017). Horizontal Whole Mount: A Novel Processing and Imaging Protocol for Thick, Three-dimensional Tissue Cross-sections of Skin. JoVE 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt BA, and Horsley V (2013). Intradermal adipocytes mediate fibroblast recruitment during skin wound healing. Development 140, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoiswohl G, Stefanovic-Racic M, Menke MN, Wills RC, Surlow BA, Basantani MK, Sitnick MT, Cai L, Yazbeck CF, Stolz DB, et al. (2015). Impact of Reduced ATGL-Mediated Adipocyte Lipolysis on Obesity-Associated Insulin Resistance and Inflammation in Male Mice. Endocrinology 156, 3610–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz O, Schürmann C, Hermes N, Müller E, Pfeilschifter J, Frank S, and Goren I (2010). Wound Healing in Mice with High-Fat Diet- or ob Gene-Induced DiabetesObesity Syndromes: A Comparative Study. Experimental Diabetes Research 2010, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JF, Sugawara A, Yamashita J, Ogura H, and Sato S (2011). Dedifferentiated fat cells: an alternative source of adult multipotent cells from the adipose tissues. Int J Oral Sci 3, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, López-Giráldez F, Dash BC, Muñoz-Rojas AR, Aultman KD, Zwick RK, Lei V, et al. (2018). Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook B, Xiao E, Kumamoto Y, Iwasaki A, and Horsley V (2016). CD301b+ Macrophages Are Essential for Effective Skin Wound Healing. J. Invest. Dermatol 136, 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn JH, Lee YK, Han JS, Jeon YG, Kim JI, Choe SS, Kim SJ, Yoo HJ, and Kim JB (2018). Perilipin 1 (Plin1) deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation. J Biol Chem 293, 13974–13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugihara H, Yonemitsu N, Miyabara S, and Yun K (1986). Primary cultures of unilocular fat cells: characteristics of growth in vitro and changes in differentiation properties. Differentiation 31, 42–49. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Takaishi S, Nagasaki M, Onozawa Y, Iino I, Maeda H, Komai T, and Oda T (2013). Medium-chain Fatty Acid-sensing Receptor, GPR84, Is a Proinflammatory Receptor. J Biol Chem 288, 10684–10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. (2013). Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Morbeck DE, Zglinicki, Von T, Van Deursen J, Lustgarten J, Scrable H, Khosla S, Jensen MD, and Kirkland JL (2010). Fat tissue, aging, and cellular senescence. Aging Cell 9, 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telerman SB, Rognoni E, Sequeira I, Pisco AO, Lichtenberger BM, Culley OJ, Viswanathan P, Driskell RR, and Watt FM (2017). Dermal Blimp1 Acts Downstream of Epidermal TGFβ and Wnt/β-Catenin to Regulate Hair Follicle Formation and Growth. J. Invest. Dermatol 137, 2270–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan GA, Hearnden CH, Oleszycka E, Lyons CL, Coutts G, O’Connell J, Corrigan MA, Lynch L, Campbell M, Callanan JJ, et al. (2014). Endogenous Oils Derived From Human Adipocytes Are Potent Adjuvants That Promote IL-1-Dependent Inflammation. Diabetes 63, 2037–2050. [DOI] [PubMed] [Google Scholar]
- Varghese M, Kimler VA, Ghazi FR, Rathore GK, Perkins GA, Ellisman MH, and Granneman JG (2019). Adipocyte lipolysis affects Perilipin 5 and cristae organization at the cardiac lipid droplet-mitochondrial interface. Sci. Rep 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboven K, Wouters K, Gaens K, Hansen D, Bijnen M, Wetzels S, Stehouwer CD, Goossens GH, Schalkwijk CG, Blaak EE, et al. (2018). Abdominal subcutaneous and visceral adipocyte size, lipolysis and inflammation relate to insulin resistance in male obese humans. Sci. Rep 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Song A, Chen W, Schwalie PC, Zhang F, Vishvanath L, Jiang L, Ye R, Shao M, Tao C, et al. (2018). Reversible De-differentiation of Mature White Adipocytes into Preadipocyte-like Precursors during Lactation. Cell Metabolism 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, and Grose R (2003). Regulation of wound healing by growth factors and cytokines. Physiol. Rev 83, 835–870. [DOI] [PubMed] [Google Scholar]
- Willenborg S, Lucas T, van Loo G, Knipper JA, Krieg T, Haase I, Brachvogel B, Hammerschmidt M, Nagy A, Ferrara N, et al. (2012). CCR2 recruits an inflammatory macrophage subpopulation critical for angiogenesis in tissue repair. Blood 120, 613–625. [DOI] [PubMed] [Google Scholar]
- Xu X, Grijalva A, Skowronski A, van Eijk M, Serlie MJ, and Ferrante AW Jr. (2013). Obesity Activates a Program of Lysosomal-Dependent Lipid Metabolism in Adipose Tissue Macrophages Independently of Classic Activation. Cell Metabolism 18, 816–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarini S, Hankin JA, Murphy RC, and Gijón MA (2014). Prostaglandins and Other Lipid Mediators. Prostaglandins and Other Lipid Mediators 113–115, 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Kumar S, Barnett AH, and Eggo MC (2000). Ceiling culture of mature human adipocytes: use in studies of adipocyte functions. J. Endocrinol 164, 119–128. [DOI] [PubMed] [Google Scholar]
- Zhang L-J, Chen SX, Guerrero-Juarez CF, Li F, Tong Y, Liang Y, Liggins M, Chen X, Chen H, Li M, et al. (2018a). Age-Related Loss of Innate Immune Antimicrobial Function of Dermal Fat Is Mediated by Transforming Growth Factor Beta. Immunity 50, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L-J, Guerrero-Juarez CF, Hata T, Bapat SP, Ramos R, Plikus MV, and Gallo RL (2015). Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science 347, 67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Di Martino JS, Bowman RL, Campbell NR, Baksh SC, Simon-Vermot T, Kim IS, Haldeman P, Mondal C, Yong-Gonzales V, et al. (2018b). Adipocyte-Derived Lipids Mediate Melanoma Progression via FATP Proteins. Cancer Discov 8, 1006–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shao M, Hepler C, Zi Z, Zhao S, An YA, Zhu Y, Ghaben AL, Wang M-Y, Li N, et al. (2019a). Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice. Journal of Clinical Investigation 129, 5327–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shao M, Hepler C, Zi Z, Zhao S, An YA, Zhu Y, Ghaben A, Wang M-Y, Li N, et al. (2019b). Dermal adipose tissue has high plasticity and undergoes reversible dedifferentiation in mice. J Clin Invest 1–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Liang H, Clarke E, Jackson C, and Xue M (2016). Inflammation in Chronic Wounds. Ijms 17, 2085–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou BO, Yu H, Yue R, Zhao Z, Rios JJ, Naveiras O, and Morrison SJ (2017a). Bone marrow adipocytes promote the regeneration of stem cells and haematopoiesis by secreting SCF. Nature Publishing Group 19, 891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P-L, Li M, Han X-W, Bi Y-H, Zhang W-G, Wu Z-Y, and Wu G (2017b). Plin5 deficiency promotes atherosclerosis progression through accelerating inflammation, apoptosis and oxidative stress. J. Cell. Biochem 1–38. [DOI] [PubMed] [Google Scholar]
- Zwick RK, Rudolph MC, Shook BA, Holtrup B, Roth E, Lei V, Van Keymeulen A, Seewaldt V, Kwei S, Wysolmerski J, et al. (2018). Adipocyte hypertrophy and lipid dynamics underlie mammary gland remodeling after lactation. Nature Communications 9, 3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Ingenuity Pathway Analysis of differentially expressed molecules in fibroblast subsets. Related to Figure 7 and Figure S7.
Table S2. Ingenuity Pathway Analysis associated diseases and functions of fibroblast subsets. Related to Figure 7 and Figure S7.
Table S3. Mouse qPCR primers. Related to STAR Methods.
Data Availability Statement
Sequencing and analysis data were deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE126514 and GSE140512.


