Abstract
Background
Antibiotic resistance (ABR) restricts the armamentarium of health-care providers against infectious diseases due to the emergence of multidrug resistance (MDR), especially in Gram-negative bacteria. This study aimed to determine pooled estimates of Gram-negative bacteria, their resistance profiles, and rates of MDR in patients with wound infection in Ethiopia.
Methods
Electronic databases such as PubMed/MEDLINE, EMBASE, Science Direct, Web of Science, and Google Scholar were searched. Original articles, available online from 1988 to 2020, addressing the prevalence and resistance patterns of Gram-negative bacteria in patients with wound infection and written in English were screened. The data were extracted using a format prepared in Microsoft Excel and exported to STATA 14.0 for the outcome analyses.
Results
The data of 15,647 wound samples, from 36 studies conducted in 5 regions of the country, were pooled. The overall pooled estimate of Gram-negative bacteria was 59% [95% CI: 52–65%, I2 = 96.41%, p < 0.001]. The pooled estimate of Escherichia colirecovered from isolates of 5205 wound samples was 17% [95% CI: 14–20%], followed by Pseudomonas aeruginosa, 11% [95% CI: 9–14%], Klebsiella pneumonia, 11% [95% CI: 9–13%], Proteus mirabilis, 8% [95% CI: 6–10%], Acinetobacter species, 4% [95% CI: 2–6%], Enterobacter species, 4% [95% CI: 3–5%], and Citrobacter species, 3% [95% CI: 2–4%]. Multidrug resistance prevalence estimates of E. coli, K. pneumonia, P. aeruginosa, P. mirabilis, Citrobacter species, Enterobacter species and Acinetobacter species were 76% [95% CI: 66–86%], 84% [95% CI: 78–91%], 66% [95% CI:43–88%], 83% [95% CI:75–91%], 87% [95% CI:78–96%], 68% [95% CI:50–87%] and 71% [95% CI:46–96%], respectively.
Conclusion
There was high resistance in Gram-negative bacteria from wound specimens to commonly used antibiotics in Ethiopia. The data warrant the need of regular epidemiological surveillance of antimicrobial resistance and implementation of an efficient infection control program.
Keywords: antibiotic resistance, Gram-negative bacteria, wound infection, meta-analysis, systematic review, Ethiopia
Introduction
Bacteriological isolation, identification, and antibiotic susceptibility testing of clinical specimens are essential tools for active surveillance of antibiotic resistance (ABR). They strongly support targeted antibiotic therapy and reduce exposure of non-involved bacterial pathogens to unnecessary antibiotics.1 Globally, ABR restricts the armamentarium of the health care providers against infectious diseases. This is mainly related to the emergence of multidrug resistant Gram-negative bacteria.2 Availability of less potent products, use of antibiotics for veterinary products, lack of standardized diagnostic facilities, and increased practice of antibiotic self-medication are directly related to the development of ABR.3–5 Worldwide, the prevalence and rate of antibiotic resistance is increasing at an alarming rate.6–8 Resistant etiologies are common causes of community and health-care associated infections.9,10 These pathogens present a concerning therapeutic challenge to clinicians with few therapeutic options left.11
To promote research and development of new antibiotics and advocate prudent use of the available ones, the World Health Organization (WHO) released a list of antibiotic-resistant priority pathogens, the majority being Gram-negative bacteria.12 These microorganisms have become a major clinical and therapeutic dilemma in the health facilities of developing countries. They are responsible for increased health-care costs owing to protracted hospital stay, morbidity and mortality.12 Multiple studies from Africa,13–15 Europe,16–19 USA,5,20–23 Australia,24–26 and Asia27–30 revealed that the huge increase of resistance in Gram-negative bacteria to available antibiotics is leading to a loss of efficacy for the treatment of many infections. Respiratory tract, urinary tract, bloodstream and wound infections are among the common conditions caused by these pathogens in health-care and community settings. Enterobacteriaceae (Klebsiella pneumonia, Escherichia coli, and Enterobacter species), Pseudomonas aeruginosa, Proteus mirabilisand Acinetobacter species have been identified as the major resistant strains responsible for these and other infections.31–34
Due to the nature of their outer layer envelope, Gram-negative bacteria are more resistant to a wide range of antibiotics than Gram-positive bacteria.35,36 A plethora of observational studies conducted across the regions of Ethiopia from various clinical settings showed increments in the prevalence of resistance patterns of Gram-negative bacteria isolates.37–43 However, there is a need of an updated pooled data set for the resistance burden of Gram-negative bacteria isolates among patients with wound infections. This might help to draw a national estimate of antibiotic resistance prevalence for these etiologies in such kinds of clinical specimens. Currently, there is one published review on the pattern of antibiotic resistance among bacteria isolated from wound samples in Ethiopia.44 Although the review was informative, its search was not exhaustive and it included only limited original studies. Moreover, the review did not estimate pooled prevalence of MDR Gram-negative bacteria. Therefore, the current review was designed to synergize the prior work by determining the pooled estimates and the resistance pattern of Gram-negative bacteria in patients with infected wounds. It also computed the rates of MDR species of Gram-negative bacteria from the available studies conducted in the country.
Methods
Reporting
This systematic review and meta-analysis was designed and reported according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement45 and meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines.46
Literature Search
The electronic databases and indexing services PubMed/MEDLINE (https://www.pubmed.ncbi.nlm.nih.gov), EMBASE (https://www.embase.com), Science Direct (https://www.sciencedirect.com), Web of Science (https://mjl.clarivate.com/search-results), and Google Scholar (https://scholar.google.com) were searched. The institutional repositories of Jimma & Addis Ababa Universities; available in Ethiopia, were also explored for grey literature. The references of all retrieved articles were checked for additional relevant publications. All searches were limited to articles written in English. The databases were searched from 1988 to March, 2020. The search terms stemmed from the following key words: prevalence, magnitude, surgical site infection, wound infections, post-operative infection, resistance, multidrug resistance, bacterial profile, antimicrobial resistance, K. pneumonia, P. aeruginosa, P. mirabilis, Citrobacter species, Enterobacter species and Acinetobacter species, Gram-negative bacteria and Ethiopia. In the advanced searching databases, the searching strategy was built based on the above-mentioned terms using the “Medical Subject Headings (MeSH)” and “All fields” by linking “AND” and “OR” Boolean operator terms as appropriate.
Study Selection and Data Extraction
Data were extracted using a standard extraction format adapted from the Joanna Briggs Institute (JBI) data extraction format.47 Two authors (LK and TM) independently reviewed the articles for data quality and methodological validity. Disagreements were resolved by consulting TAM. A pre-designed data collection format was used to extract data from each included study. The extracted information includes the name of the first author, year of publication, study period, study design, study region, age of patients (years), gender, total sample size, the number of Gram-negative bacteria species and antibiotic resistance patterns for E. coli, K. pneumonia, P. aeruginosa, P. mirabilis, Enterobacter species, Citrobacter species and Acinetobacter species. References and data for each study were carefully cross-checked to avoid duplication of data and maintain the integrity of the meta-analysis. All retrieved studies were exported to Mendeley desktop reference manager to organize search outcomes and for removal of duplicate articles.
Risk of Bias and Quality Assessment
The quality assessment for the included studies was based on the JBI study quality appraisal criteria established for cross-sectional and cohort studies. The JBI’s appraisal criteria consist of nine questions. The first three questions measure the appropriateness of the sampling frame. These questions address the target population, describing appropriateness of sampling method and adequacy of sample size. The next three questions assess the study subjects and setting. The last three questions measure the appropriateness of the statistical analysis and adequacy of the response rate. Since all the studies that fulfill the eligibility criteria of this systematic review and meta-analysis had scored 50% and above, none of them had poor quality status and all of them were considered.
Eligibility Criteria
The studies were selected based on predefined inclusion criteria. The published and unpublished studies reporting the epidemiology of Gram-negative bacteria and their ABR profile from 1988 to March 2020 in patients with wound infection, according to the criteria of the Clinical Laboratory Standards Institute (CLSI),48 were included. All the included studies were published in English and conducted in Ethiopia. Studies conducted on non-clinical specimens such as foods, food handlers’ belongings, health workers’ belongings, health workers’ carriage or animals and of non-infectious carriage were excluded. Studies without full-text, qualitative studies, reviews, commentaries, case series, case reports, conference proceedings and abstracts were also excluded.
Data Processing and Analysis
Data were extracted in Microsoft Excel format and analyzed using STATA 14.0 statistical software. The prevalence data for ABR were pooled using “”metaprop” command. Random effect model was applied to estimate the pooled prevalence and antibiotic resistance pattern of Gram-negative bacteria species in patients with wound infection. A potential source of heterogeneity was investigated by subgroup and meta-regression analysis. The existence of heterogeneity among studies was examined by I2 heterogeneity test. The I2 values of 0, 25, 50 and 75% were considered as no, low, moderate and high heterogeneities, respectively.49–51 Thus, the DerSimonian–Laird random effects model was employed.52 The geographic distribution (regional states), and year of publication were considered for subgroup analysis to minimize the random variations between the point estimates of the primary study. The Begg’s funnel plot and Egger’s regression test were used to evaluate the possibility of publication bias. Forest plot format was used to present the pooled prevalence with 95% confidence interval (CI). Two-sided p values < 0.05 were considered as statistically significant.
Outcome Measurement
The three major outcomes reported in this systematic review and meta-analysis were the pooled prevalence estimates, the rate of antibiotic resistance isolates and the rate of multidrug resistance of Gram-negative bacteria isolates in patients with infected wounds in Ethiopia. The pooled prevalence estimates of common Gram-negative bacteria species were the number of each Gram-negative bacteria isolates divided by the number of all the detected isolates. The pooled estimates of resistance to each tested antibiotic was calculated by dividing the number of resistance isolates of each species by the total number of all the detected isolates of the species. Multidrug resistance was defined as resistance to at least two antibiotics by the isolated Gram-negative bacteria for the various antibiotics.
Results
Characteristics of the Included Studies
Of 987 identified studies, 497 were duplicates and were excluded upon reviewing the titles and abstracts. Further, 392 studies were excluded because they were irrelevant. Then, 98 full articles were assessed for eligibility. Of these, 62 articles were excluded, as irrelevant (N =56) and review articles (N= 6). Finally, 36 studies meeting the inclusion were included in this review (Figure 1).
Figure 1.
Preferred Reporting Items for Systematic review and Meta-Analysis (PRISMA) flow chart for the included studies.
All studies were conducted from April 1983 to September 2018 and published online from 1988 to 2020. Of the 36 included studies, 32 were cross sectional and 4 were cohort studies. All but 6 studies were unpublished, which were obtained from Addis Ababa and Jimma University repositories. Over two-thirds of the studies were conducted in Amhara region (n = 15) followed by Addis Ababa city (n = 11), Oromia region (n = 5), SNNPR (n = 3), and Tigray region (n = 2). The age of the study participants in the included studies ranged from 1 month to 100 years and the sample size ranged from 50 to 1627. Table 1 summarizes the study characteristics and the number of Gram-negative (E. coli, P. aeruginosa, K. pneumonia, P. mirabilis, Enterobacter species, Citrobacter species, and Acinetobacter species) isolates recovered from wound samples.
Table 1.
The Study Characteristics, Quality, and the Number of Gram-Negative Isolates Recovered from Wound Samples
| Name of First Author | Year of Publication | Study Period | Study Design | Regional State | Age | Gender | Sample Size |
E. coli (n) |
K. pneumonia (n) |
P. aeruginosa (n) |
P. mirabilis (n) |
EB. Species (n) |
CB. Species (n) |
Acinetobacter Species (n) |
Overall JBI’s Score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | |||||||||||||||
| Godebo53 | 2013 | June to Dec 2011 | Cross sectional | Oromia | NR | NR | NR | 384 | 30 | 46 | 74 | 107 | 18 | 7 | 8 | |
| Mama54 | 2014 | May to September 2013 | Cross sectional | Oromia | 6 months–90 years | 107 | 43 | 150 | 29 | 14 | 11 | 23 | 9 | |||
| Mengesha55 | 2014 | January to June 2012 | Cross sectional | Tigray | 15–79 years | 467 | 143 | 610 | 6 | 29 | 15 | 15 | 4 | 8 | ||
| Feleke56 | 2018 | February 1 to May 31, 2016 | Cross sectional | Amhara | 1–85 years | NR | NR | 116 | 24 | 29 | - | - | 8 | |||
| Ali57 | 2018 | May to September, 2016 | Cross sectional | Oromia | 34.12 ± 16.86 years | 168 | 282 | 450 | 14 | 5 | - | - | 8 | |||
| Dessie58 | 2016 | October 2013 to March 2014 | Cross sectional | Addis Ababa | 8–80 years | NR | NR | 1088 | 22 | 10 | 6 | 1 | 3 | 23 | 6 | |
| Tesfahunegn59 | 2009 | November 2005 and April 2006 | Cross sectional | Tigray | 35.5 years | 134 | 112 | 246 | 14 | 7 | 1 | 1 | 1 | 7 | ||
| Yallew60 | 2016 | March to July 2015 | Cross sectional | Amhara | 16–40 years | NR | NR | 908 | 22 | 30 | 25 | 8 | 17 | 6 | 6 | 6 |
| Mulu61 | 2012 | October 2010 to January 2011 | Cross sectional | Amhara | 32.2 years | 96 | 198 | 294 | 9 | 2 | 5 | 4 | 8 | |||
| Kalayu62 | 2020 | April 2016 to April 2017 | Cross sectional | Addis A baba | 9 months–88 years | 285 | 364 | 649 | 11 | 10 | - | 1 | 2 | 1 | 3 | 8 |
| Asres63 | 2017 | March to August 2015 | Cross sectional | Addis Ababa | 0–85 years | 118 | 79 | 179 | 24 | 15 | 8 | 1 | 2 | 17 | 8 | |
| Sahile64 | 2016 | June, 2012 to February; 2013 | Cross sectional | Oromia | 18–75 years | 103 | 97 | 200 | 9 | 5 | 8 | 7 | 4 | 6 | 4 | 6 |
| Amare65 | 2011 | January 2010 and June 2010 | Cross sectional | Amhara | 7–75 years | NR | NR | 1627 | 27 | 6 | 3 | 4 | 11 | 6 | ||
| Endalafer66 | 2010 | 2007 and April 2008 | Cross sectional | Addis Ababa | 17–79 years | 130 | 85 | 215 | 16 | 12 | 12 | - | 7 | |||
| Tekie67 | 2008 | April 2006 to July 2006 | Cross sectional | Addis Ababa | 1–80 years | 97 | 76 | 173 | 7 | 4 | 10 | 2 | 2 | 2 | 9 | |
| Kifilie68 | 2019 | January 2016 to May 2016 | Cross sectional | Amhara | 15–44 years | 0 | 107 | 107 | 20 | 14 | 2 | 1 | 4 | 4 | 8 | |
| Dessalegn69 | 2013 | November 2010 to March 2011 | Cross sectional | SNNPR | 12–100 years | 116 | 78 | 194 | 45 | 24 | 12 | 18 | 7 | |||
| Misha70 | 2020 | April 20 to August 20, 2019 | Cohort | Oromia | 38 ± 16.30 years | 125 | 126 | 251 | 8 | 5 | 7 | 6 | 4 | 8 | ||
| Gelaw71 | 2014 | November 2010 to February 2011 | Cross sectional | Amhara | NR | 27 | 15 | 510 | 10 | 11 | 8 | 5 | 4 | 2 | 7 | |
| Ali72 | 2016 | July 22 – October 25, 2016 | Cross sectional | Amhara | 16–76 years | 87 | 251 | 338 | 9 | 7 | 2 | 1 | 3 | 8 | ||
| Melaku73 | 2012 | April-August 2009 | Cohort | Amhara | NR | NR | NR | 1383 | 49 | 36 | 26 | 6 | ||||
| Bitew74 | 2019 | January 1 to May 31, 2016 | Cross sectional | Amhara | 15–44 years | 0 | 222 | 222 | 24 | - | 1 | 1 | 1 | 1 | 6 | |
| Biadglegne75 | 2009 | September 2003 to June 2008 | Cross sectional | Amhara | 3months-90 years | 222 | 157 | 397 | 4 | 10 | 9 | 17 | 7 | 2 | 6 | |
| Bitew76 | 2018 | June 2016 to July 2017 | Cross sectional | Addis Ababa | 15–64 years | 213 | 153 | 366 | 49 | 12 | 14 | 7 | 6 | 4 | 4 | 8 |
| Hailu77 | 2016 | 1 January 2013 to 30 December 2015 | Cross sectional | Amhara | 4 months−76 years | 195 | 185 | 380 | 33 | 20 | 26 | 22 | 5 | 5 | 7 | |
| Abraham78 | 2009 | November, 2007 and May, 2008 | Cross sectional | Addis Ababa | 4–75 years | 158 | 33 | 191 | 17 | 12 | 16 | 6 | 7 | |||
| Mulu79 | 2006 | - | Cross sectional | Amhara | NR | NR | NR | 151 | 8 | 7 | - | 3 | 7 | |||
| Lema80 | 2012 | August 2006 to May 2007 | Cross sectional | Addis Ababa | 13 to 92 years | 157 | 88 | 245 | 14 | 2 | 7 | 47 | 7 | |||
| Ayalew81 | 2014 | December 2013 to May 2014 | Cross sectional | Addis Ababa | 10–68 years | 268 | 32 | 300 | 19 | 21 | 53 | 24 | 9 | 8 | 9 | |
| Mohammed82 | 2017 | March to May, 2014 | Cross sectional | Amhara | 2–80 years | 81 | 56 | 137 | 15 | 17 | 16 | 13 | 13 | 15 | 2 | 7 |
| Habte-Gabr83 | 1988 | April 1983 and January 1984 | Cohort | Addis Ababa | NR | 733 | 273 | 1006 | 54 | 46 | 40 | 94 | 14 | 14 | 1 | 6 |
| Billoro84 | 2019 | January 1 to September 1, 2017 | Cohort | SNNPR | NR | 147 | 108 | 355 | 5 | 25 | 5 | 7 | ||||
| Guta85 | 2014 | November 2010 to June 2011 | Cross sectional | SNNPR | NR | NR | NR | 100 | 30 | 32 | 16 | 12 | 3 | 4 | 7 | |
| Azene86 | 2011 | - | Cross sectional | Amhara | 1–90 years | 368 | 231 | 599 | 82 | 12 | 92 | 55 | 21 | 21 | 8 | |
| Sewunet87 | 2013 | April to July 2010 | Cross sectional | Addis Ababa | 7–55 years | 30 | 20 | 50 | - | 2 | 15 | 4 | 7 | |||
| Motbainor88 | 2020 | April 1 to July 31, 2018 | Cross sectional | Amhara | - | 129 | 109 | 238 | - | - | 11 | - | 9 | |||
Pooled Estimates of Gram-Negative Bacteria Isolates from Wound Samples
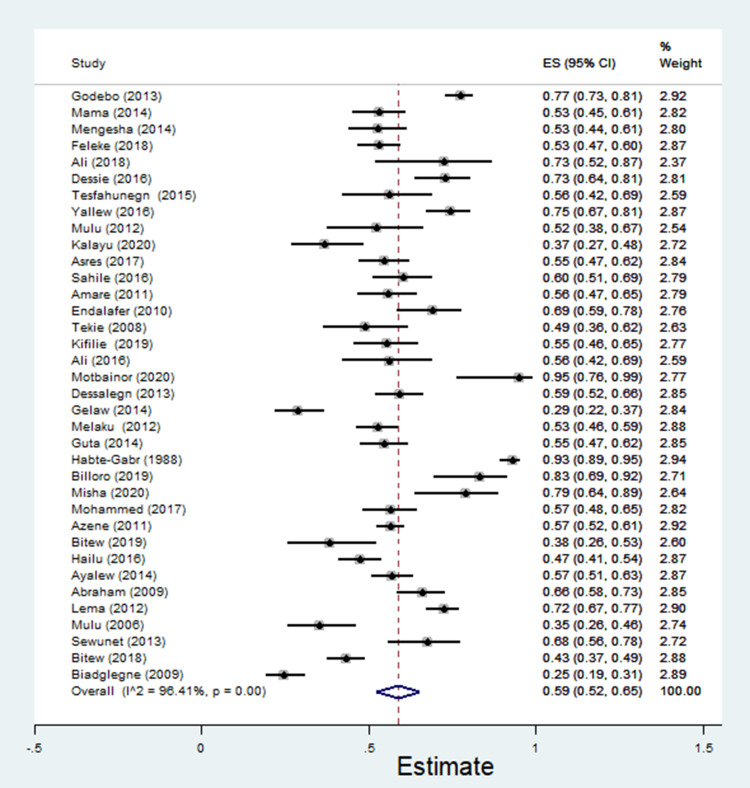
A total of 5,376 bacterial isolates, the majority (n = 3150, 58.6%) being Gram-negative isolates, were recovered from 15,647 wound samples. In the random-effects model, the pooled estimate of Gram-negative bacteria was 59% [95% CI: 52–65%, I2 = 96.41%, χ2 = 973.93, p < 0.001, Figure 2] with substantial heterogeneity. The estimates of common Gram-negative bacteria isolated from wound samples are described in Table 2.
Figure 2.
The pooled estimates of Gram-negative bacteria isolates from wound samples.
Table 2.
The Estimates of Common Gram-Negative Bacteria Isolated from Wound Samples
| Bacteria | No. of Studies | Sample | Case | Estimate [95% CI] | χ2 | I2 (%) |
|---|---|---|---|---|---|---|
| Escherichia coli | 34 | 5205 | 762 | 17[14–20] | 380.24 | 91.32 |
| K. pneumoniae | 34 | 5281 | 539 | 11[9–13] | 364.42 | 90.94 |
| P. aeruginosa | 32 | 6145 | 562 | 11[8–11] | 557.33 | 94.44 |
| P. mirabilis | 30 | 4713 | 513 | 8[6–10] | 369.37 | 92.15 |
| Citrobacter species | 20 | 3563 | 126 | 3[2–4] | 39.81 | 52.27 |
| Enterobacter species | 16 | 2857 | 123 | 4[3–5] | 29.41 | 49.00 |
| Acinetobacter species | 12 | 1786 | 80 | 4[2–6] | 72.86 | 84.90 |
Abbreviation: CI, confidence interval.
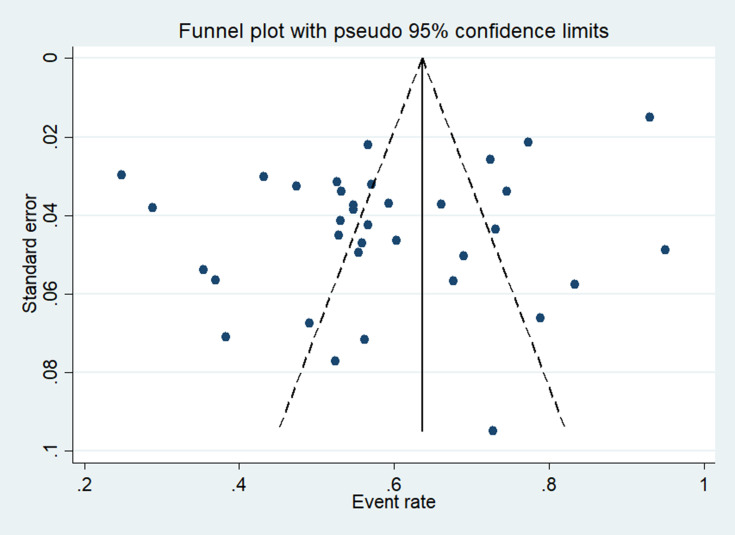
The symmetry of the funnel plot visual inspection of standard error with prevalence of Gram-negative bacteria indicated the absence of publication bias. This finding was statistically confirmed by Egger’s regression test (p = 0.12 and Begg’s test, p = 0.87) [Figure 3].
Figure 3.
The funnel plot for the publication bias of the included studies for the prevalence of Gram-negative bacteria.
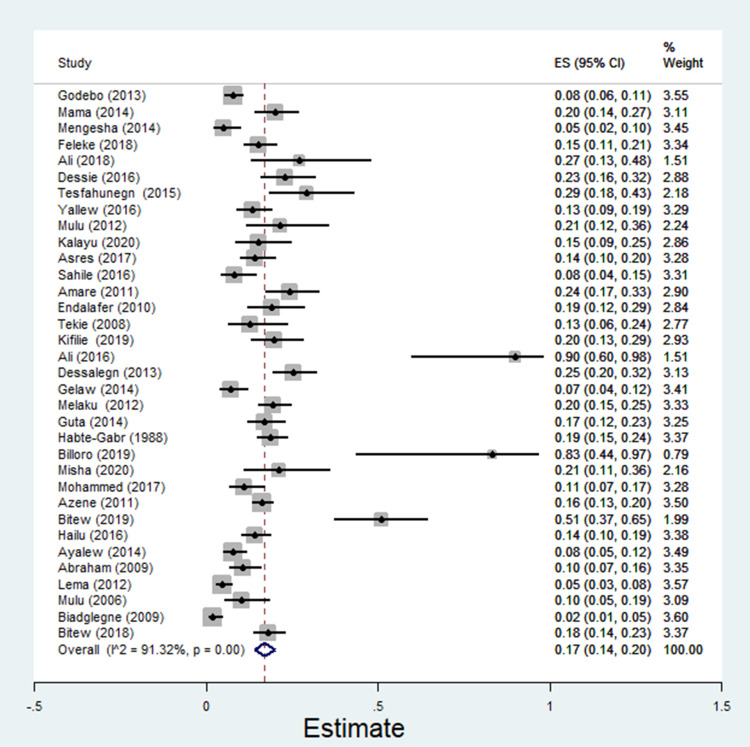
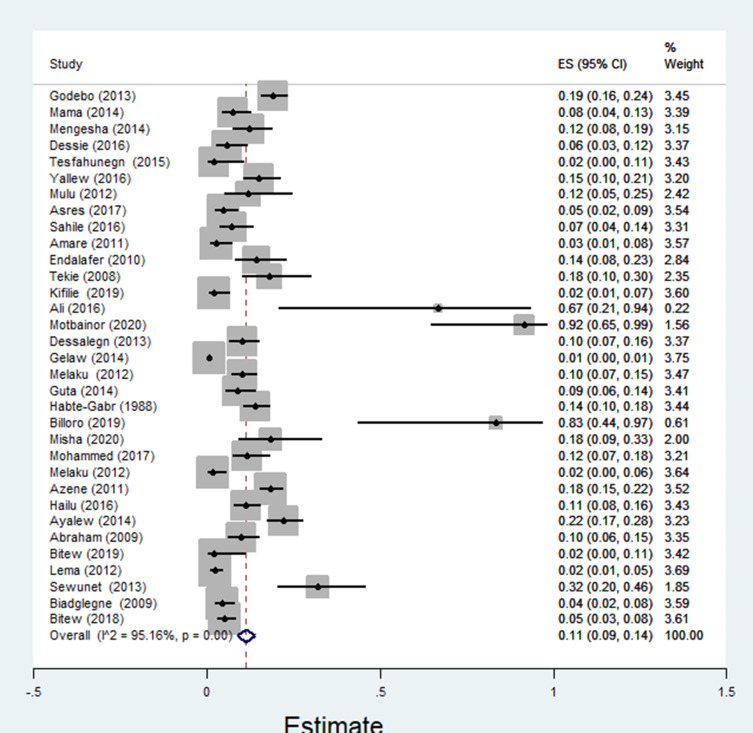
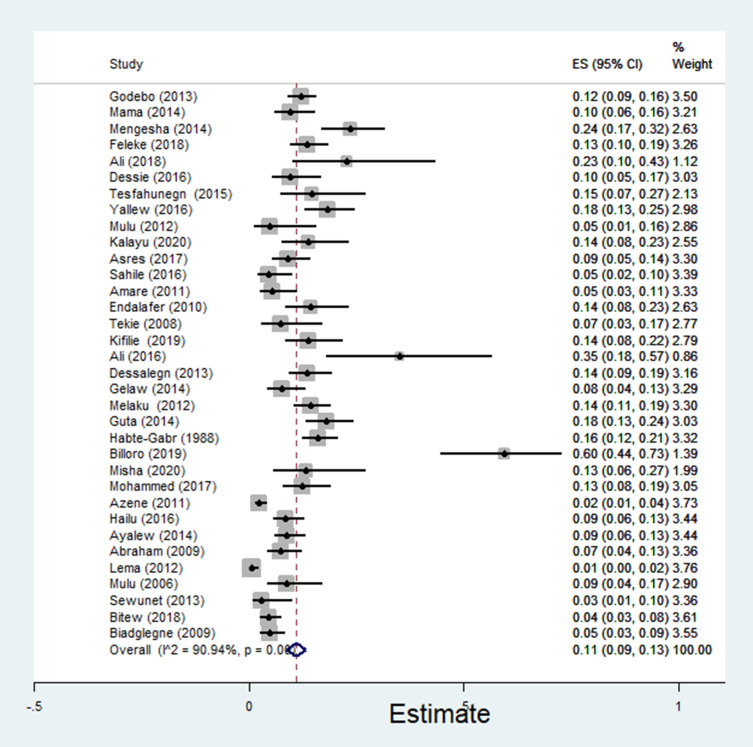
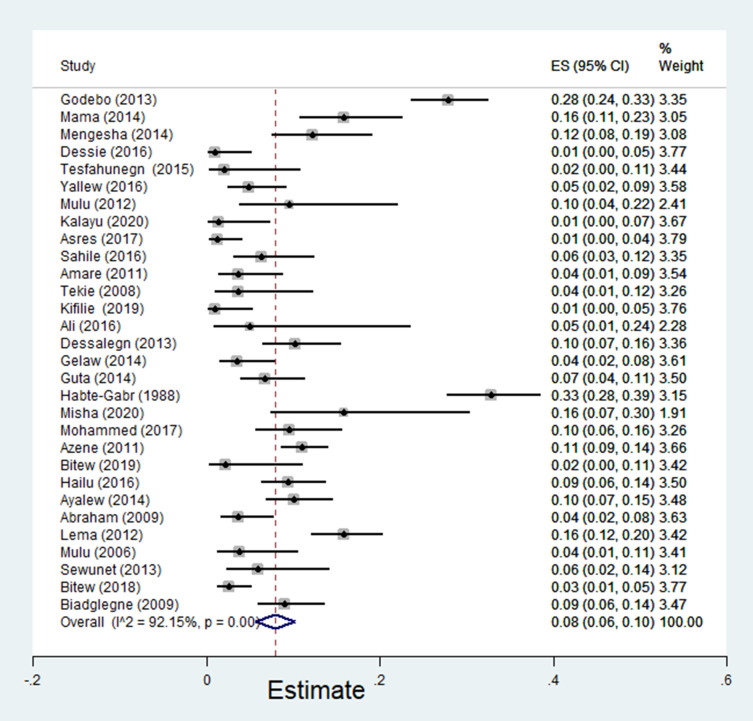
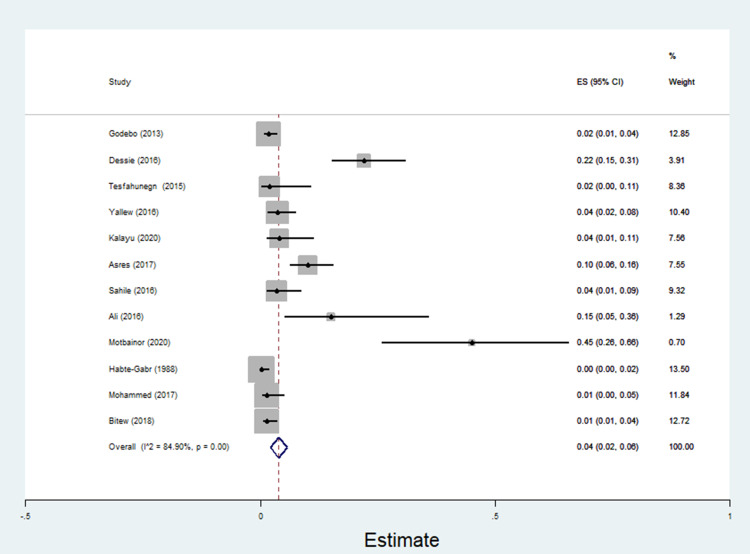
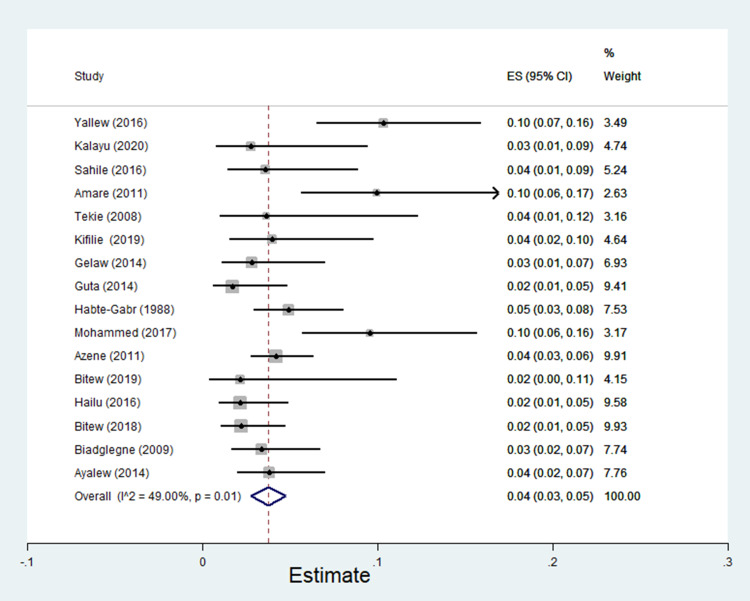
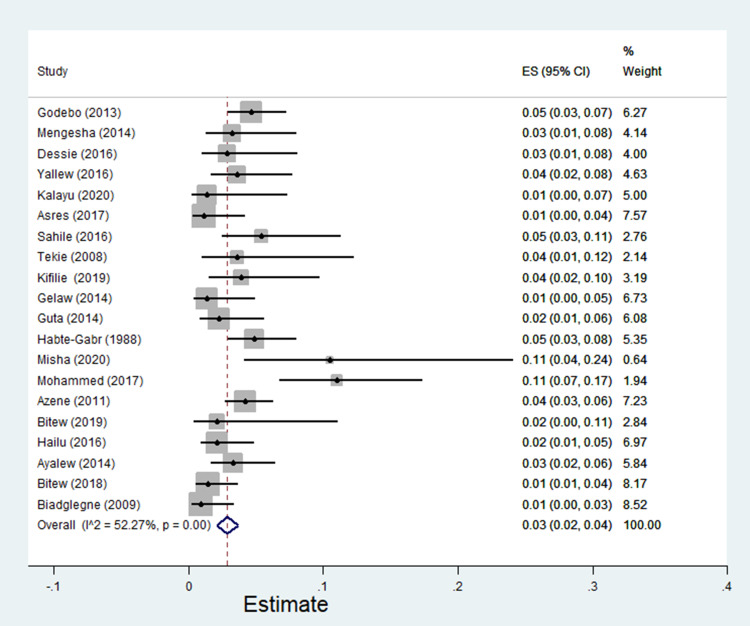
The pooled estimate of E. coli isolates recovered from 5205 wound samples was 17% [95% CI: 14–20%, Figure 4]. Similarly, the pooled estimate of K. pneumonia was 11% [95% CI: 9–13%, Figure 5], P. aeruginosa, 11% [95% CI: 8–13%, Figure 6], P. mirabilis, 8% [95% CI: 6–10%, Figure 7], Citrobacter species, 3% [95% CI: 2–4%, Figure 8], Enterobacter species, 4% [95% CI: 3–5%, Figure 9] and Acinetobacter species, 4% [95% CI: 2–6%, Figure 10].
Figure 4.
Forest plot showing pooled estimate of E. coli bacteria among patients with wound infection.
Figure 5.
Forest plot showing pooled estimate of Klebsiella pneumoniae among patients with wound infection.
Figure 6.
Forest plot showing pooled estimate of P. aeruginosa among patients with wound infection.
Figure 7.
Forest plot showing pooled estimate of Proteus mirabilis among patients with wound infection.
Figure 8.
Forest plot showing pooled estimate of Citrobacter species among patients with wound infection.
Figure 9.
Forest plot showing pooled estimate of Enterobacter species among patients with wound infection.
Figure 10.
Forest plot showing pooled estimate of Acinetobacter species among patients with wound infection.
Antibiotic Resistance Patterns of Gram-Negative Bacteria Isolates
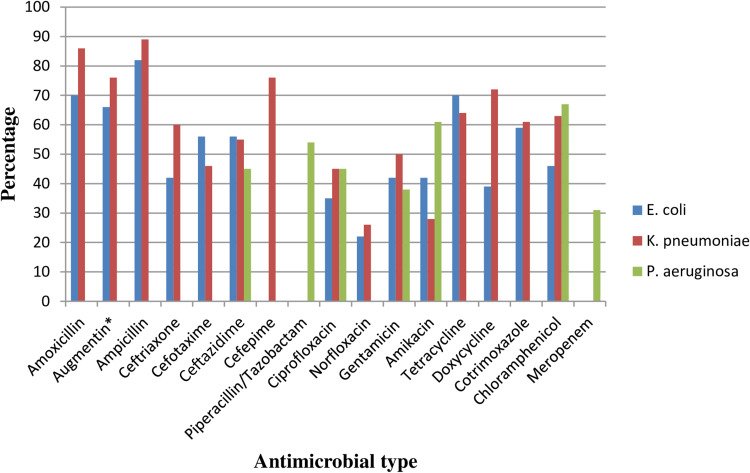
This finding indicated the Gram-negative bacteria E. coli exhibited the highest resistance to ampicillin, 82% [95% CI: 76–88%]. Isolates of E. coli were also resistant to amoxicillin, 79% [95% CI: 55–85%], tetracycline, 70% [955 CI: 61–78%], doxycycline, 39% [95% CI: 0–78%], amoxicillin-clavulanic acid, 66% [95% CI: 52–81%], cotrimoxazole, 59% [95% CI: 48–69%], ceftazidime, 56% [95% CI: 25–86%], cefotaxime, 56% [95% CI: 21–90%], ceftriaxone, 42% [95% CI: 30–53%], chloramphenicol, 46% [95% CI: 12–72%], amikacin 42% [95% CI: 12–72%] and gentamicin, 42% [95% CI: 24–61%]. Relatively lower rates of resistance were observed to norfloxacin, 22% [95% CI: 10–35%] and ciprofloxacin, 35% [95% CI: 17–53%] among E. coli isolates.
Similarly, the highest pooled estimates of resistance among K. pneumonia isolates were observed to ampicillin, 89% [95 CI: 85–93%], amoxicillin, 86% [95% CI: 80–93%], amoxicillin-clavulanic acid, 76% [95% CI: 64–88%], cefepime, 76% [95% CI: 56–92%], cefotaxime, 46% [95% CI: 19–72%], and ciprofloxacin, 45% [95% CI: 28–61%]. However, the lowest resistance profile of K. pneumonia was observed to norfloxacin, 26% [95% CI: 15–38%] and amikacin, 28% [95% CI: 7–48%]. The highest rate of resistance among P. aeruginosa isolates was associated with chloramphenicol, 67% [95% CI: 54–81%], amikacin, 61% [95% CI: 4–100%], and piperacillin, 54% [95% CI: 5–100%], but meropenem, 31% [95% CI: 12–50%] tended to have the highest barrier to resistance [Table 3 and Figure 11].
Table 3.
The Pooled Estimates of Antimicrobial Resistance Among Gram-Negative Bacteria Isolates of Wound Samples
| Antimicrobials | Pooled Estimates of Resistant Isolates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K. pneumonia | P. aeruginosa | |||||||||||||
| No. of Studies | Sample Size | Case | ES [95% CI] | I2 (%) | No. of Studies | Sample Size | Case | Estimate [95% CI] | I2 (%) | No. of Studies | Sample Size | Case | ES [95% CI] | I2 (%) | |
| Amoxicillin | 14 | 335 | 204 | 70 [55–85] | 92.59 | 11 | 184 | 153 | 86[80–93] | 48.43 | ND | ND | ND | ND | ND |
| Augmentin* | 12 | 281 | 182 | 66[52–81] | 89.53 | 11 | 178 | 126 | 76[64–88] | 78.27 | ND | ND | ND | ND | ND |
| Ampicillin | 20 | 415 | 334 | 82 [76–88] | 78.01 | 18 | 266 | 227 | 89[85–93] | 00.00 | ND | ND | ND | ND | ND |
| Ceftriaxone | 20 | 546 | 216 | 42 [30–53] | 90.94 | 19 | 322 | 181 | 60[48–73] | 86.44 | ND | ND | ND | ND | ND |
| Cefotaxime | 4 | 97 | 57 | 56[21–90] | 94.61 | 6 | 111 | 44 | 46[19–72] | 91.50 | ND | ND | ND | ND | ND |
| Ceftazidime | 5 | 128 | 59 | 56[25–86] | 94.02 | 6 | 66 | 35 | 55[31–79] | 82.10 | 7 | 77 | 43 | 45[11–79] | 93.28 |
| Cefepime | ND | ND | ND | ND | ND | 2 | 22 | 16 | 76[56–92] | -NS | ND | ND | ND | ND | ND |
| Piperacillin/Tazobactam | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 4 | 61 | 38 | 54[5–100] | 98.01 |
| Ciprofloxacin | 19 | 503 | 174 | 35 [17–53] | 97.06 | 17 | 270 | 113 | 45[28–61] | 91.81 | 17 | 318 | 148 | 45[24–67] | 96.77 |
| Norfloxacin | 8 | 143 | 34 | 22[10–35] | 81.97 | 8 | 152 | 42 | 26[15–38] | 63.92 | ND | ND | ND | ND | ND |
| Gentamicin | 20 | 504 | 244 | 42[24–61] | 97.56 | 22 | 349 | 164 | 50[36–64] | 91.19 | 23 | 443 | 192 | 38[17–59] | 97.98 |
| Amikacin | 5 | 124 | 46 | 42[12–72] | 95.06 | 3 | 55 | 13 | 28[7–48] | 66.08 | 3 | 39 | 29 | 61[4–100] | 94.12 |
| Tetracycline | 17 | 372 | 266 | 70[61–78] | 72.40 | 14 | 213 | 133 | 64[50–77] | 83.79 | ND | ND | ND | ND | ND |
| Doxycycline | 4 | 157 | 36 | 39[0–78] | 97.56 | 4 | 74 | 54 | 72[52–92] | 71.76 | ND | ND | ND | ND | ND |
| Cotrimoxazole | 21 | 524 | 318 | 59[48–69] | 88.82 | 17 | 274 | 171 | 61[48–75] | 88.52 | ND | ND | ND | ND | ND |
| Chloramphenicol | 19 | 495 | 246 | 46[32–60] | 92.73 | 18 | 270 | 167 | 63[51–75] | 81.01 | 16 | 369 | 254 | 67[54–81] | 90.04 |
| Meropenem | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | 2 | 21 | 7 | 31[12–50] | - |
| P. mirabilis | Enterobacter Species | Citrobacter Species | |||||||||||||
| No. of studies | Sample size | Case | ES [95% CI] | I2 (%) | No of studies | Sample size | case | Estimate [95% CI] | I2 (%) | No of studies | Sample size | case | ES [95% CI] | I2 (%) | |
| Amoxicillin | 7 | 161 | 98 | 73[54–92] | 88.90 | 5 | 48 | 25 | 63[27–99] | 88.92 | 7 | 48 | 25 | 60[41–80] | 48.14 |
| Augmentin* | 8 | 122 | 69 | 59[37–82] | 86.84 | 3 | 13 | 8 | 65[40–89] | 00.00 | 4 | 16 | 9 | 58[34–81] | 00.00 |
| Ampicillin | 16 | 322 | 249 | 81[74–89] | 69.02 | 8 | 57 | 48 | 87[79–96] | 9 | 60 | 51 | 90[83–97] | 00.00 | |
| Ceftriaxone | 15 | 316 | 111 | 49[33–66] | 93.29 | 7 | 66 | 25 | 42[24–59] | 58.74 | 8 | 76 | 27 | 34[21–47] | 32.01 |
| Cefotaxime | 5 | 146 | 24 | 30[11–49] | 86.50 | 2 | 15 | 4 | 26[4–48] | – | 4 | 32 | 10 | 30[15–46] | 00.00 |
| Ciprofloxacin | 13 | 326 | 95 | 40[18–61] | 96.85 | 7 | 55 | 30 | 52[30–75] | 67.87 | 5 | 60 | 23 | 38[9–67] | 84.63 |
| Norfloxacin | 7 | 224 | 27 | 16[7–26] | 75.52 | 2 | 7 | 5 | 72[39–100] | – | 3 | 28 | 6 | 23[0–45] | 40.77 |
| Gentamicin | 18 | 389 | 158 | 38[18–57] | 96.40 | 11 | 85 | 51 | 55[31–78] | 86.19 | 7 | 74 | 41 | 53[31–75] | 73.84 |
| Amikacin | 2 | 26 | 10 | 38[20–57] | – | 2 | 16 | 11 | 70[48–92] | – | ND | ND | ND | ND | ND |
| Tetracycline | 11 | 213 | 167 | 76[66–86] | 66.93 | 8 | 75 | 50 | 66[50–83] | 61.94 | 8 | 64 | 42 | 67[53–80] | 23.83 |
| Doxycycline | 4 | 189 | 98 | 49[7–91] | 97.78 | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| Cotrimoxazole | 14 | 354 | 209 | 54[40–67] | 84.74 | 10 | 82 | 51 | 58[39–77] | 74.54 | 8 | 76 | 53 | 61[37–84] | 86.29 |
| Chloramphenicol | 13 | 329 | 217 | 59[46–72] | 82.09 | 7 | 70 | 46 | 66[47–85] | 72.51 | 6 | 70 | 51 | 72[54–90] | 69.26 |
| Acinetobacter species | |||||||||||||||
| Antimicrobials | No of studies | Sample size | case | Estimate [95% CI] | I2 (%) | ||||||||||
| Amoxicillin-clavulanic acid | 5 | 122 | 47 | 85[76-94] | 00.00 | ||||||||||
| Ampicillin | 7 | 52 | 43 | 85[74-97] | 22.58 | ||||||||||
| Ceftriaxone | 7 | 65 | 51 | 81[69-93] | 32.54 | ||||||||||
| Cefotaxime | 5 | 59 | 48 | 85[74-96] | 30.15 | ||||||||||
| Ciprofloxacin | 9 | 72 | 44 | 57[39-75] | 63.68 | ||||||||||
| Norfloxacin | 2 | 11 | 3 | 16[1-45] | - | ||||||||||
| Gentamicin | 8 | 69 | 44 | 68[56-79] | 12.01 | ||||||||||
| Tetracycline | 6 | 59 | 38 | 70[50-90] | 65.87 | ||||||||||
| Cotrimoxazole | 6 | 44 | 34 | 89[68-93] | 16.32 | ||||||||||
| Chloramphenicol | 5 | 52 | 37 | 72[60-84] | 00.00 | ||||||||||
| Cefazidime | 6 | 59 | 50 | 88[80-96] | 00.00 | ||||||||||
| Cefepime | 2 | 6 | 3 | 50[12-88] | - | ||||||||||
| Piperacillin-Tazobactam | 2 | 12 | 10 | 86[67-100] | - | ||||||||||
Figure 11.
Percentage of E. coli, K pneumonia, and P. aeruginosa resistant to different antibiotics commonly in use in Ethiopian settings.
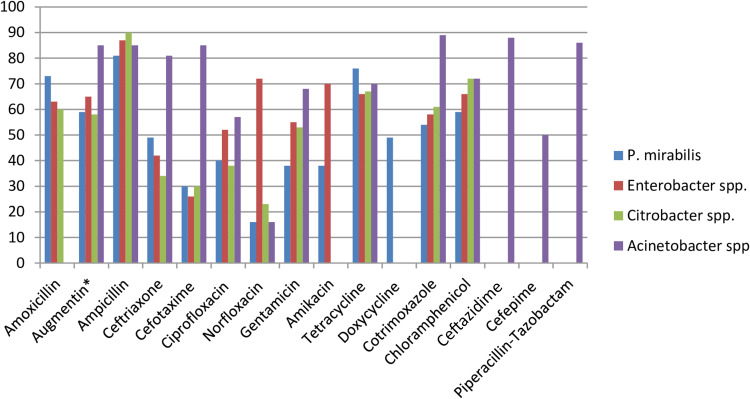
Among P. mirabilis isolates, higher rates of resistance were reported to ampicillin, 81% [95% CI: 74–89%], tetracycline, 76% [95% CI: 66–86%], amoxicillin, 73% [95% CI: 54–92%], amoxicillin-clavulanic acid, 59% [95% CI: 37–82%], and chloramphenicol, 59% [95% CI: 46–72%]. However, the isolates appeared relatively sensitive to norfloxacin, 16% [95% CI: 7–26%], cefotaxime, 30% [95% CI: 11–49%], amikacin, 38% [95% CI: 20–57%], and gentamicin, 38% [95% CI: 18–57%]. Although Enterobacter species were fairly sensitive to cefotaxime, 26% [95% CI: 4–48%] and ceftriaxone, 42% [95% CI: 24–59%], a significant proportion of isolates were resistant to ampicillin, 87% [95% CI: 79–96%], norfloxacin, 72% [95% CI: 39–100%], and amikacin, 70% [95% CI: 48–92%]. Besides, the lowest pooled rate of resistance to norfloxacin, 23% [95% CI: 0–45%], cefotaxime, 30% [95% CI: 15–46%], and ceftriaxone, 34% [95% CI: 21–47%] was observed among Citrobacter species. Resistance rates of Acinetobacter species were also reported by 12 studies consisting of 1786 samples. High levels of resistance were observed to cotrimoxazole 89% [95% CI: 68–93%], ceftazidime, 88% [95% CI: 80–96%], piperacillin, 86% [95% CI: 67–100%], cefotaxime, 85% [955 CI: 74–96%], and ampicillin, 85% [95% CI: 74–97%], while norfloxacin, 16% [95% CI: 1–45%] tended to have a more effective barrier to resistance in Acinetobacter species [Table 3 and Figure 12].
Figure 12.
Percentage of P. mirabilis, Enterobacter species, Citrobacter species, and Acinetobacter species resistant to different antibiotics commonly in use in Ethiopian settings.
Patterns of Multidrug Resistance Among Gram-Negative Bacteria Isolates
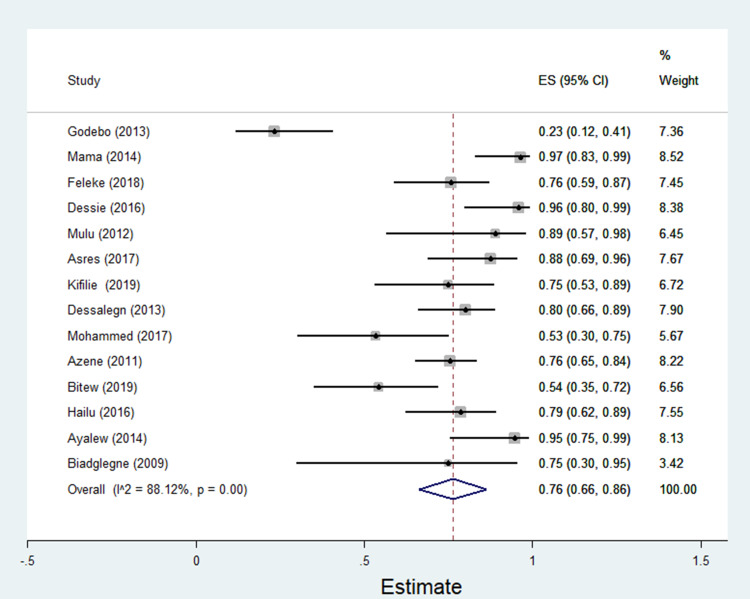
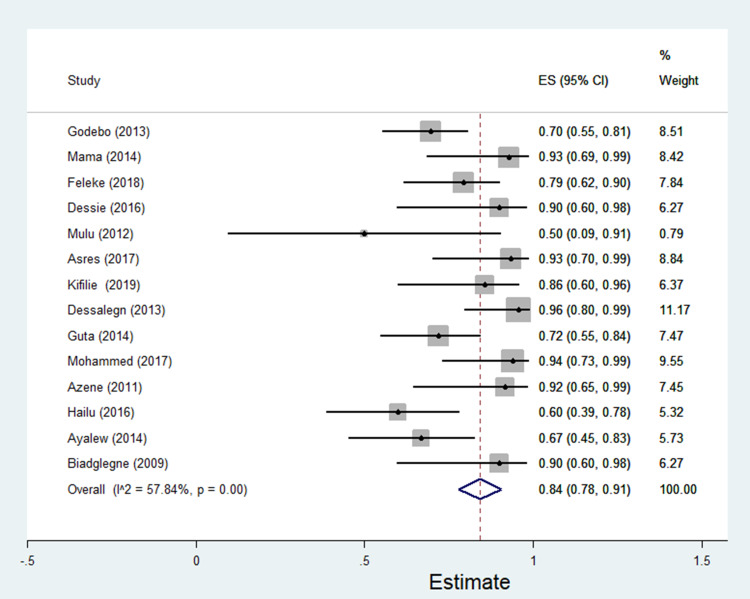
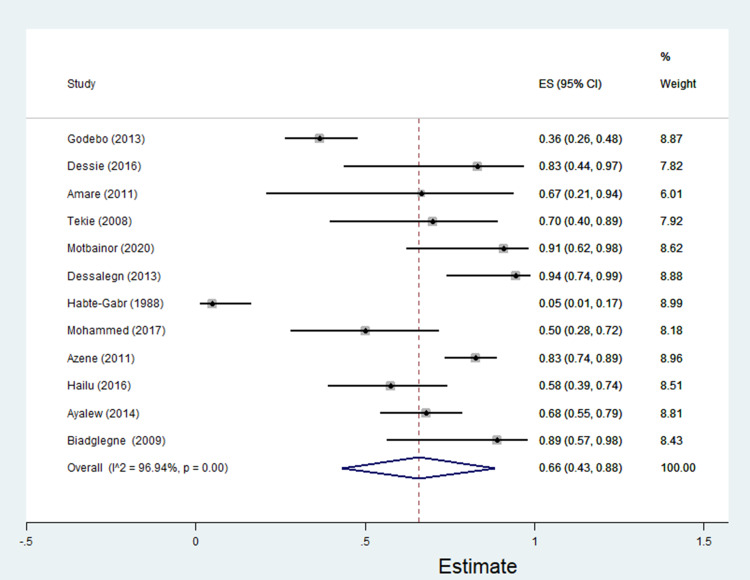
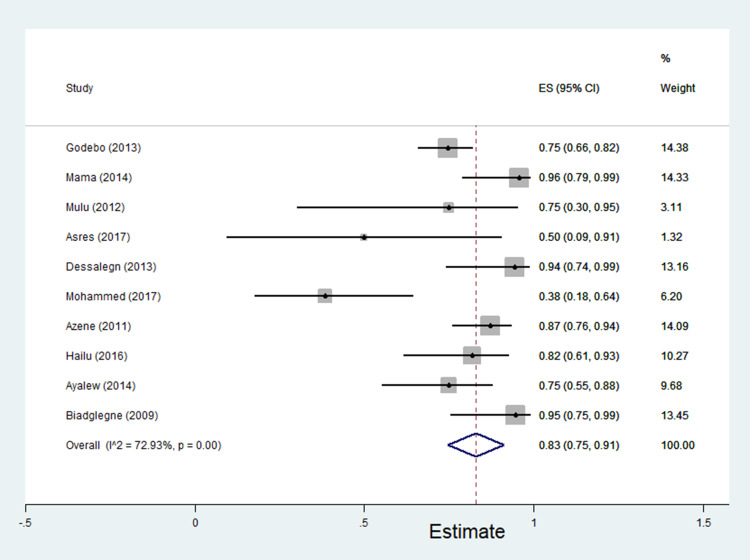
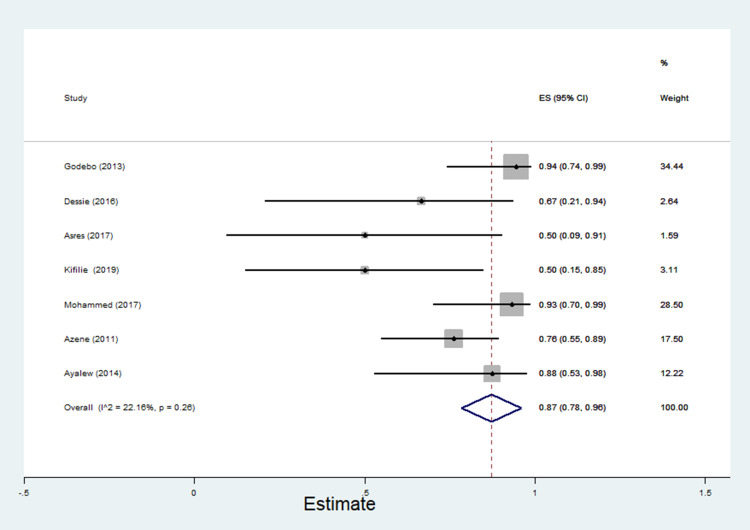
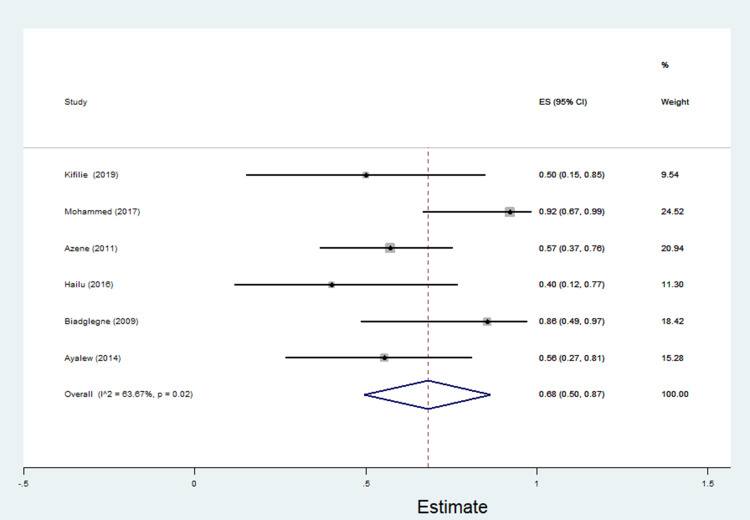
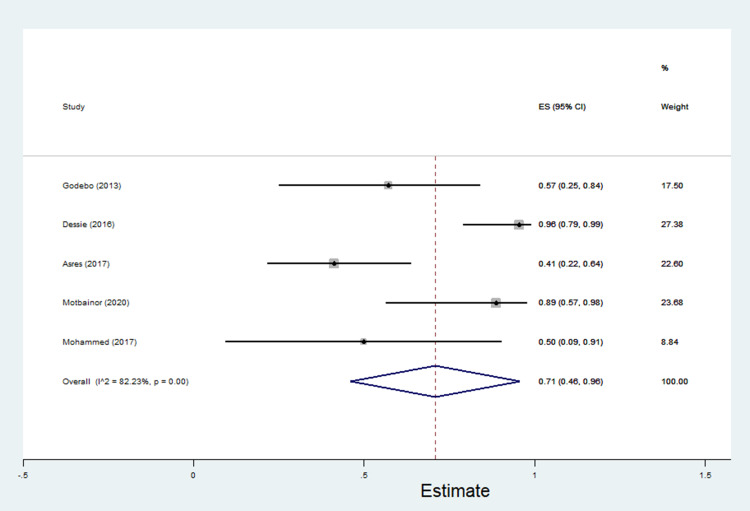
The data of multidrug resistance profiles of Gram-negative bacteria for the various antibiotics tested in the included studies was analyzed. Accordingly, the pooled estimate of MDR in E. coli was 76% [95% CI: 66–86%], (Figure 13). The pooled estimates of MDR in K. pneumonia, P. aeruginosa, P. mirabilis, Citrobacter species, Enterobacter species, and Acinetobacter species were 84% [95% CI: 78–91%], (Figure 14), 66% [95% CI:43–88%], (Figure 15), 83%[95% CI: 75–91%], (Figure 16), 87% [95% CI: 78–96%], (Figure 17), 68%[95% CI:50–87%], (Figure 18) and 71%[95% CI: 46–96%], (Figure 19), respectively.
Figure 13.
Percentage of multidrug resistance in E. coli to different antimicrobials commonly in use in Ethiopia.
Figure 14.
Percentage of multidrug resistance in K. pneumonia to different antimicrobials commonly in use in Ethiopia.
Figure 15.
Percentage of multidrug resistance in P. aeruginosa to different antimicrobials commonly in use in Ethiopia.
Figure 16.
Percentage of multidrug resistance in P. mirabilis to different antimicrobials commonly in use in Ethiopia.
Figure 17.
Percentage of multidrug resistance in Enterobacter species to different antimicrobials commonly in use in Ethiopia.
Figure 18.
Percentage of multidrug resistance in Citrobacter species to different antimicrobials commonly in use in Ethiopia.
Figure 19.
Percentage of multidrug resistance in Acinetobacter species to different antimicrobials commonly in use in Ethiopia.
Therefore, it is prudent to observe that more than two-thirds of the Gram-negative bacteria isolates recovered from wound samples were resistant to at least two antibiotics i.e. had multidrug resistance. Table 4 depicts patterns of MDR Gram-negative bacteria isolates recovered from wound samples.
Table 4.
Pooled Estimates of Multidrug Resistance Among Gram-Negative Bacteria Isolates of Wound Samples
| Bacteria | Pooled Estimates of Multidrug Resistant Isolates | ||||
|---|---|---|---|---|---|
| No. of Studies | Sample Size | Case | Estimate [95% CI] | I2 (%) | |
| E. coli | 14 | 391 | 293 | 76[66–86] | 88.12 |
| K. pneumoniae | 14 | 266 | 212 | 84[78–91] | 57.84 |
| P. aeruginosa | 12 | 358 | 213 | 66[43–88] | 96.94 |
| P. mirabilis | 10 | 287 | 230 | 83[75–91] | 72.93 |
| Citrobacter species | 7 | 71 | 59 | 87[78–96] | 22.16 |
| Enterobacter species | 6 | 59 | 39 | 68[50–87] | 63.67 |
| Acinetobacter species | 5 | 58 | 42 | 71[46–96] | 82.23 |
Abbreviation: CI, confidence interval.
Discussions
Antibiotic resistance is a natural process which occurs when bacteria evolve to resist the medicines that are being used to combat them. It is one of the greatest tragedies of the 21st century, which has undermined progress in health care, food production, and life expectancy.89
This review summarized the epidemiology of Gram-negative bacteria and their antibiotic-resistance pattern in patients with wound infection in Ethiopia. Accordingly, the rate of wound infection by Gram-negative bacteria was 59%. This finding is relatively lower than a study in Tanzania where Gram-negative bacteria accounted for 85.2% of the cases.15 It was noted that the pooled estimates of E. coli (17%), K. pneumonia (11%), P. aeruginosa (11%), P. mirabilis (8%), Acinetobacter species (4%), Citrobacter species (4%), and Enterobacter species (3%) is in agreement with a study conducted in Tanzania where the prevalence of P. mirabilis (16%) and P. aeruginosa (13%) was higher in wound infections than in those with no wound infection (9% and 6%, respectively).15 Moreover, a previous review in Ethiopia also reported that the pooled estimates of E. coli, K. pneumonia, P. aeruginosa, and P. mirabilis were 13%, 9%, 9%, and 8%, respectively.44
Therefore, E. coli, K. pneumonia, P. aeruginosa, P. mirabilis, Citrobacter species, Enterobacter species, and Acinetobacter species were the most prevalent Gram-negative pathogens with an alarming rate of resistance to commonly used antibiotics. The rates of antibiotic resistance among important Gram-negative pathogens are increasing.90,91 This study showed the prevalence of drug resistance among E. coli, K. pneumonia, P. aeruginosa, P. mirabilis, Enterobacter species, and Acinetobacter species has exceeded 50% for penicillin. More than 40% of the strains were also resistant to third-generation cephalosporin and aminoglycosides. These pathogens are commonly implicated in both community and nosocomial infections.92 Lucas et al.93 reported 50–100% resistance to ampicillin and cotrimoxazole, 20–47% to gentamicin, and 46–69% to ceftriaxone among strains of K. pneumonia and E. coli. In other studies, nosocomial Gram-negative pathogens such as E. coli, K. pneumonia, and P. aeruginosa are becoming increasingly resistant to commonly used antimicrobial agents such as third-generation cephalosporin and penicillin.92,94 These antibiotics, the vital lifelines that are the backbone of health-care systems in low- and middle-income countries, are becoming obsolete due to the emerging threat of antibiotic resistance. The adverse outcomes of this disaster are huge and concerning. It is estimated to cost more than 700,000 lives annually worldwide and the number is expected to grow to 10 million by the year 2050.95 If no action is taken, the lost global production between now and 2050 would be an enormous 100 trillion USD.95 In low- and low-middle-income countries, it will be more tragic as the already crippled health systems will be further compromised due to limited antibiotics choices and unaffordable treatment costs.96
A large pool of evidence shows that the reckless use of antibiotics plays a pivotal role in the advent of multidrug resistant bacteria.97–99 According to this review, Citrobacter species (87%), K. pneumonia (84%), P. mirabilis (83%), E. coli (76%), Acinetobacter species (71%), Enterobacter species (68%), and P. aeruginosa (66%) were the most recognized multidrug resistant bacteria in Ethiopian settings. Lim et al. found K. pneumoniae, Acinetobacter species, E. coli, P. aeruginosa as the most common multidrug resistant bacteria posing a significant threat to the health-care system.100 Similarly, E. coli, K. pneumonia, and P. aeruginosa are identified as the common multidrug resistant Gram-negative bacilli inflicting an intolerable harm to global health. They are associated with substantial morbidity and mortality, and increased health-care costs.101 Furthermore, E. coli, K. pneumonia, P. aeruginosa, and Acinetobacter species are resistant to almost all currently available antibiotics.102
The essence of epidemiological surveillance of bacterial infection and bacterial resistance to existing antibiotics is to create awareness and strengthen the implementation of infection prevention and control (IPC) strategies.103 Given the frequent polymicrobial nature of wound infection, a compiled data set of bacteriological investigations is required to demonstrate the burden of resistant pathogens in sub-Saharan Africa, a region which lacks data on the true extent of the problem.92 This is also indispensable for resource-limited settings such as Ethiopia, where the health-care facilities have a rudimentary antimicrobial stewardship and poor IPC activities.103 Available antimicrobial resistance data will sensitize clinicians and policymakers. These data have a grave importance for revising the existing national treatment protocols to guide optimal antimicrobial therapy. These data also contribute to combat antimicrobial drug resistance and re-direct the habit of antimicrobial prescription in health-care facilities. Over time, all bacteria will acquire mechanisms of resistance to current and future antibiotics. This is a harsh fact that will continue to become a deadly reality to the planet. Therefore, this is a critical time which needs the initiation of programs to curtail antibiotic resistance and the development of newer antibiotic agents.
Strength and Limitations of the Study
Although this review addresses an important evidence gap through identifying and synthesizing data about the prevalence of Gram-negative bacteria species, patterns of antibiotic resistance and multidrug resistance among patients with wound infection in Ethiopia, the authors acknowledge that this review has some limitations. The major limitations were the issue of heterogeneity and representativeness. The study period, hospital settings, population characteristics, methods of bacterial identification, and antibiotic susceptibility tests were also varied across the studies questioning the appropriateness of combining the study findings to create a single pooled estimate. While the conclusion of high antibiotic resistance among Gram-negative bacteria in patients with wound infection was drawn from the study findings, the original studies included in the analyses were only from four regions and one city out of the nine regions and two cities of the country. Hence, the representativeness of the results to the remaining regions and city of the country might be questioned. Moreover, the reader should note that the protocol of this review was not published online ahead of the actual meta-analysis.
However, this review provides useful information about the current status of antibiotic resistance among Gram-negative bacteria in wound infection. Despite the considerable variations in the location of the study, sample size and duration across studies which contribute to the differences between groups, the current review has paramount importance in providing a full picture of the problem at national level to help policymakers to design cost-effective control and treatment strategies. Besides, the findings could help the concerned bodies to pay attention to further research in the areas where there is paucity of data or even no published study.
Conclusion
Resistance among Gram-negative organisms is widespread to commonly used antibiotics in Ethiopian patients with infected wounds. These data warrant the need for regular epidemiological surveillance of antibiotic resistance and implementation of an efficient infection control and stewardship program. Future research efforts should also focus on the transmission dynamics and enhanced generation and aggregation of the solitary data. The need for research and development to enrich the pipeline for novel antimicrobial compounds is a matter of survival. This will invigorate the hope of mankind on the planet and an excellent tool to counteract the threat posed by antibacterial resistance.
Funding Statement
The authors received no specific funding for this work.
Abbreviations
ABR, antibiotic resistance; GNB, Gram-negative bacteria; CI, confidence interval; MDR, multi-drug resistance; ; SNNPR, Southern Nation, Nationalities, and People’s Region; STATA, South Texas Art Therapy Association.
Data Sharing Statement
All relevant data are within the manuscript.
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Author Contributions
LC, TM, and TAM conducted the database search, screening, and quality assessment. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Vernet G, Mary C, Altmann DM, et al. Surveillance for antimicrobial drug resistance in under-resourced countries. Emerg Infect Dis. 2014;20(3):434–441. doi: 10.3201/EID2003.121157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Márió G, Zoltán B, Marianna Á, Andrea L, Katalin B. Characterization of resistance in gram-negative urinary isolates using existing and novel indicators of clinical relevance: a 10-year data analysis. Life. 2020;10(16):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila J. Tibor Pal: Update on Antibacterial Resistance in Low-Income Countries: Factors Favoring the Emergence of Resistance. Open Infect Dis. 2010;4:38–54. [Google Scholar]
- 4.Larissa G, George G, Roger Z, Shivanki J, Jean L. Use of antibiotics without a prescription in the U S population: a scoping review. Ann Intern Med. 2019;171:257–263. [DOI] [PubMed] [Google Scholar]
- 5.Adeel A, Márió G, Che S, et al. Evidence of the practice of self-medication with antibiotics among the lay public in low- and middle-income countries: a scoping review. Antibiotics. 2020;9(597):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, Abramson MA, Beekmann SE, et al. Antimicrobial resistance among Gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J Clin Microbiol. 2007;45(10):3352–3359. doi: 10.1128/JCM.01284-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Souli M, Galani I, Giamarellou H. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Eurosurveillance. 2008;13(47):1–11. [PubMed] [Google Scholar]
- 8.Ko W-C, Hsueh P-R. Increasing extended-spectrum β-lactamase production and quinolone resistance among Gram-negative bacilli causing intra-abdominal infections in the Asia/Pacific region: data from the Smart Study 2002–2006. J Infect. 2009;59(2):95–103. doi: 10.1016/j.jinf.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 9.Huttner A, Harbarth S, Carlet J, et al. Antimicrobial resistance: a global view from the 2013 World Healthcare-Associated Infections Forum. Antimicrob Resist Infect Control. 2013;2(31):1–13. doi: 10.1186/2047-2994-2-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tacconelli E, Karchmer AW, Yokoe D, D’Agata EM. Preventing the influx of vancomycin-resistant enterococci into health care institutions, by use of a simple validated prediction rule. Clin Infect Dis. 2004;39(7):964–970. doi: 10.1086/423961 [DOI] [PubMed] [Google Scholar]
- 11.Márió G, Katalin B, Gabriella T. esistance levels and epidemiology of non-fermenting gram-negative bacteria in urinary tract infections of inpatients and outpatients (renfuti): a10-year epidemiological snapshot. Antibiotics. 2019;8(143):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Organization WH. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. World Health Organization; 3–88. 2017. [Google Scholar]
- 13.Singh N, Manchanda V. Control of multidrug-resistant gram-negative bacteria in low-and middle-income countries—high impact interventions without much resources. Clin Microbiol Infect. 2017;23(4):216–218. doi: 10.1016/j.cmi.2017.02.034 [DOI] [PubMed] [Google Scholar]
- 14.Le Doare K, Bielicki J, Heath PT, Sharland M. Systematic review of antibiotic resistance rates among gram-negative bacteria in children with sepsis in resource-limited countries. J Pediatric Infect Dis Soc. 2015;4(1):11–20. doi: 10.1093/jpids/piu014 [DOI] [PubMed] [Google Scholar]
- 15.Carroll M, Rangaiahagari A, Musabeyezu E, Singer D, Ogbuagu O. Five-year antimicrobial susceptibility trends among bacterial isolates from a tertiary health-care facility in Kigali, Rwanda. Am J Trop Med Hyg. 2016;95(6):1277–1283. doi: 10.4269/ajtmh.16-0392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumburu HH, Sonda T, Mmbaga BT, et al. Patterns of infections, aetiological agents and antimicrobial resistance at a tertiary care hospital in northern Tanzania. Trop Med Int Health. 2017;22(4):454–464. doi: 10.1111/tmi.12836 [DOI] [PubMed] [Google Scholar]
- 17.Kaier K, Wilson C, Chalkley M, et al. Health and economic impacts of antibiotic resistance in European hospitals-outlook on the BURDEN project. Infection. 2008;36(5):492–494. doi: 10.1007/s15010-008-7453-0 [DOI] [PubMed] [Google Scholar]
- 18.Nellums LB, Thompson H, Holmes A, et al. Antimicrobial resistance among migrants in Europe: a systematic review and meta-analysis. Lancet Infect Dis. 2018;18(7):796–811. doi: 10.1016/S1473-3099(18)30219-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Kraker ME, Davey PG, Grundmann H, Group BS. Mortality and hospital stay associated with resistant Staphylococcus aureus and Escherichia coli bacteremia: estimating the burden of antibiotic resistance in Europe. PLoS Med. 2011;8(10):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawkey P. The growing burden of antimicrobial resistance. J Antimicrob Chemother. 2008;62(suppl_1):i1–i9. doi: 10.1093/jac/dkn241 [DOI] [PubMed] [Google Scholar]
- 21.Critchley IA, Cotroneo N, Pucci MJ, Mendes R. The burden of antimicrobial resistance among urinary tract isolates of Escherichia coli in the United States in 2017. PLoS One. 2019;14(12):1–11. doi: 10.1371/journal.pone.0220265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sahm DF, Marsilio MK, Piazza G. Antimicrobial resistance in key bloodstream bacterial isolates: electronic surveillance with the Surveillance Network Database—USA. Clin Infect Dis. 1999;29(2):259–263. doi: 10.1086/520195 [DOI] [PubMed] [Google Scholar]
- 23.Diekema D, Beach M, Pfaller M, Jones R, SP G. Antimicrobial resistance in viridans group streptococci among patients with and without the diagnosis of cancer in the USA, Canada and Latin America. Clin Microbiol Infect. 2001;7(3):152–157. doi: 10.1046/j.1198-743x.2001.00230.x [DOI] [PubMed] [Google Scholar]
- 24.Styers D, Sheehan DJ, Hogan P, Sahm DF. Laboratory-based surveillance of current antimicrobial resistance patterns and trends among Staphylococcus aureus: 2005 status in the United States. Ann Clin Microbiol Antimicrob. 2006;5(2):1–9. doi: 10.1186/1476-0711-5-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wozniak TM, Paterson D, Halton K. Review of the epidemiological data regarding antimicrobial resistance in Gram-negative bacteria in Australia. Infect Dis Health. 2017;22(4):210–218. doi: 10.1016/j.idh.2017.07.003 [DOI] [Google Scholar]
- 26.Wozniak TM, Bailey EJ, Graves N. Health and economic burden of antimicrobial-resistant infections in Australian hospitals: a population-based model. Infect Control Hosp Epidemiol. 2019;40(3):320–327. doi: 10.1017/ice.2019.2 [DOI] [PubMed] [Google Scholar]
- 27.Campbell AJ, Daley DA, Bell JM, et al. Progress towards a coordinated, national paediatric antimicrobial resistance surveillance programme: staphylococcus aureus, enterococcal and Gram-negative bacteraemia in Australia. J Antimicrob Chemother. 2020;75(6):1639–1644. doi: 10.1093/jac/dkaa065 [DOI] [PubMed] [Google Scholar]
- 28.Ria B, Márió G, Mária M, et al. Prevalence and antibiotic resistance of ESKAPE pathogens isolated in the emergency department of a tertiary care teaching hospital in hungary: a 5-year retrospective survey. Antibiotics. 2020;9(624):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jean -S-S, Hsueh P-R. High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents. 2011;37(4):291–295. doi: 10.1016/j.ijantimicag.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 30.Lai -C-C, Lee K, Xiao Y, et al. High burden of antimicrobial drug resistance in Asia. J Global Antimicrob Resist. 2014;2(3):141–147. doi: 10.1016/j.jgar.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 31.Kang C-I, Song J-H. Antimicrobial resistance in Asia: current epidemiology and clinical implications. Infect Chemother. 2013;45(1):22–31. doi: 10.3947/ic.2013.45.1.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaurasia S, Sivanandan S, Agarwal R, Ellis S, Sharland M, Sankar MJ. Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ. 2019;364:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Angelis G, D’Inzeo T, Fiori B, Spanu T, Sganga G. Burden of antibiotic resistant Gram negative bacterial infections: evidence and limits. J Med Microbiol Diagn. 2014;3(1):1–6. [Google Scholar]
- 34.Rossolini GM, Mantengoli E, Docquier J, Musmanno RA, Coratza G. Epidemiology of infections caused by multiresistant Gram-negatives: eSBLs, MBLs, panresistant strains. Microbiologica-bologna-. 2007;30(3):332–339. [PubMed] [Google Scholar]
- 35.Oduro-Mensah D, Obeng-Nkrumah N, Bonney EY, et al. Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Ann Clin Microbiol Antimicrob. 2016;15(29):1–9. doi: 10.1186/s12941-016-0144-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Márió G. The concept of an ideal antibiotic: implications for drug design: review. Molecules. 2019;24(892):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Exner M, Bhattacharya S, Christiansen B, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hygiene Infect Control. 2017;12:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zenebe T, Kannan S, Yilma D, Beyene G. Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma University specialized hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci. 2011;21(1):1–8. doi: 10.4314/ejhs.v21i1.69038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennet R, Eriksson M, Tafari N, Nord CE. Intestinal bacteria of newborn Ethiopian infants in relation to antibiotic treatment and colonisation by potentially pathogenic gram-negative bacteria. Scand J Infect Dis. 1991;23(1):63–69. doi: 10.3109/00365549109023376 [DOI] [PubMed] [Google Scholar]
- 40.Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):1–13. doi: 10.1371/journal.pone.0222911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulu W, Abera B, Yimer M, Hailu T, Ayele H, Abate D. Bacterial agents and antibiotic resistance profiles of infections from different sites that occurred among patients at Debre Markos Referral Hospital, Ethiopia: a cross-sectional study. BMC Res Notes. 2017;10(254):1–9. doi: 10.1186/s13104-016-2345-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mamuye Y. Antibiotic resistance patterns of common Gram-negative uropathogens in St. Paul’s Hospital Millennium Medical College Ethiopian Journal of Health Sciences. 2016;26(2):93–100. doi: 10.4314/ejhs.v26i2.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alemayehu T, Ali M, Mitiku E, Hailemariam M. The burden of antimicrobial resistance at tertiary care hospital, southern Ethiopia: a three years’ retrospective study. BMC Infect Dis. 2019;19(1):1–8. doi: 10.1186/s12879-019-4210-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sisay M, Worku T, Edessa D. Microbial epidemiology and antimicrobial resistance patterns of wound infection in Ethiopia: a meta-analysis of laboratory-based cross-sectional studies. BMC Pharmacol Toxicol. 2019;20(35):1–19. doi: 10.1186/s40360-019-0315-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med. 2015;162(11):777–784. doi: 10.7326/M14-2385 [DOI] [PubMed] [Google Scholar]
- 46.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 47.Peters MD, Godfrey CM, McInerney P, Soares CB, Khalil H, Parker D. The Joanna Briggs Institute reviewers’ manual 2015: methodology for JBI scoping reviews. 2015. [Google Scholar]
- 48.Coyle MB. Manual of Antimicrobial Susceptibility Testing. BCIT Imaging Services; 2005. [Google Scholar]
- 49.Rücker G, Schwarzer G, Carpenter JR, Schumacher M. Undue reliance on I2 in assessing heterogeneity may mislead. BMC Med Res Methodol. 2008;8(79):1–9. doi: 10.1186/1471-2288-8-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ioannidis JP. Interpretation of tests of heterogeneity and bias in meta‐analysis. J Eval Clin Pract. 2008;14(5):951–957. doi: 10.1111/j.1365-2753.2008.00986.x [DOI] [PubMed] [Google Scholar]
- 51.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta‐analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 52.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 53.Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12(7):1–7. doi: 10.1186/1476-0711-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13(14):1–10. doi: 10.1186/1476-0711-13-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mengesha RE, Kasa BG-S, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder Teaching and Referral Hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7(1):1–6. doi: 10.1186/1756-0500-7-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feleke T, Eshetie S, Dagnew M, et al. Multidrug-resistant bacterial isolates from patients suspected of nosocomial infections at the University of Gondar Comprehensive Specialized Hospital, Northwest Ethiopia. BMC Res Notes. 2018;11(602):1–7. doi: 10.1186/s13104-018-3709-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ali S, Birhane M, Bekele S, et al. Healthcare associated infection and its risk factors among patients admitted to a tertiary hospital in Ethiopia: longitudinal study. Antimicrob Resist Infect Control. 2018;7(2):1–9. doi: 10.1186/s13756-017-0298-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dessie W, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. Int J Microbiol. 2016;2016:1–8. doi: 10.1155/2016/2418902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeamanuel T, Asrat D, Yimtubezinash W, Kidanu E. Bacteriology of surgical site and catheter related urinary tract infections among patients admitted in Mekelle Hospital, Mekelle, Tigray, Ethiopia. Ethiop Med J. 2009;47(2):117–127. [PubMed] [Google Scholar]
- 60.Yallew WW, Kumie A, Yehuala FM. Point prevalence of hospital-acquired infections in two teaching hospitals of Amhara region in Ethiopia. Drug Healthc Patient Saf. 2016;8:71–76. doi: 10.2147/DHPS.S107344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mulu W, Kibru G, Beyene G, Damtie M. Postoperative nosocomial infections and antimicrobial resistance pattern of bacteria isolates among patients admitted at Felege Hiwot Referral Hospital, Bahirdar, Ethiopia. Ethiop J Health Sci. 2012;22(1):7–18. [PMC free article] [PubMed] [Google Scholar]
- 62.Kalayu AA, Di Riba K, Girma C, Abdella E. Incidence and bacterial etiologies of surgical site infections in a Public Hospital, Addis Ababa, Ethiopia. Open Microbiol J. 2019;13:301–307. doi: 10.2174/1874285801913010301 [DOI] [Google Scholar]
- 63.Asres G, Legese M, Woldearegay G. Prevalence of multidrug resistant Bacteria in postoperative wound infections at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Arch Med. 2017;9(4):1–9. [Google Scholar]
- 64.Sahile T, Esseye S, Beyene G, Ali S. Post-surgical infection and antibiotic susceptibility patterns of bacteria isolated from admitted patients with signs of infection at Jimma University specialized hospital, Jimma, Ethiopia. Int J Trop Dis Health. 2016;17(4):1–12. doi: 10.9734/IJTDH/2016/27253 [DOI] [Google Scholar]
- 65.Amare B, Abdurrahman Z, Moges B, et al. Postoperative surgical site bacterial infections and drug susceptibility patterns at Gondar University Teaching Hospital, Northwest Ethiopia. J Bacteriol Parasitol. 2011;2(8):1–6. doi: 10.4172/2155-9597.1000126 [DOI] [Google Scholar]
- 66.Endalafer N. Bacterial Nosocomial Infections and Their Antimicrobial Susceptibility Patterns in Surgical Wards and Surgical Intensive Care Unit of Tikur Anbessa University Hospital. Addis Ababa; 2008. http://etd.aau.edu.et/handle/123456789/6067. [Google Scholar]
- 67.Ekie K Surgical wound infection in Tikur Anbessa hospital with special emphasis on Pseudomonas aeruginosa. Unpublished MSc thesis in medical microbiology, Addis Ababa University, Medical Faculty, Ethiopia; Available from: http://etdaaueduet/bitstream/handle/123456789/5772/Kassaye%20Tekiepdf [Google Scholar]
- 68.Bitew Kifilie A, Dagnew M, Tegenie B, Yeshitela B, Howe R, Abate E. Bacterial profile, antibacterial resistance pattern, and associated factors from women attending postnatal health service at university of Gondar teaching hospital, northwest ethiopia. Int J Microbiol. 2018;2018:1–11. doi: 10.1155/2018/3165391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S. Aerobic bacterial isolates from post-surgical wound and their antimicrobial susceptibility pattern: a hospital based cross-sectional study. J Med Res. 2014;3(2):18–23. [Google Scholar]
- 70.Gemedo Misha LC, Melaku T. Incidence, risk factors, antimicrobial susceptibility patterns and outcomes of surgical site infections among patients admitted to Jimma Medical Center, South West Ethiopia: prospective Cohort Study. Unpublished. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gelaw A, Gebre-Selassie S, Tiruneh M, Mathios E, Yifru S. Isolation of bacterial pathogens from patients with postoperative surgical site infections and possible sources of infections at the University of Gondar Hospital, Northwest Ethiopia. J Environ Occupational Health. 2014;3(2):103–108. [Google Scholar]
- 72.Abdurrahman Ali KD. Incidence of Surgical Site Infection, Predisposing Factors and Associated Costs at Dessie Referral Hospital, Dessie, Ethiopia. 2017. [Google Scholar]
- 73.Melaku S, Gebre-Selassie S, Damtie M, Alamrew K. Hospital acquired infections among surgical, gynaecology and obstetrics patients in Felege-Hiwot referral hospital, Bahir Dar, northwest Ethiopia. Ethiop Med J. 2012;50(2):135–144. [PubMed] [Google Scholar]
- 74.Bitew AM, Tegene BM, Yeshitila BM, Howe R, Dagnew M. Bacterial profile, antibacterial susceptibility pattern and associated factors among women attending antenatal and post-natal health services at the university of Gondar teaching hospital, northwest Ethiopia. Ethiop Med J. 2019;S1:10–22. [Google Scholar]
- 75.Biadglegne F, Abera B, Alem A, Anagaw B. Bacterial isolates from wound infection and their antimicrobial susceptibility pattern in Felege Hiwot referral Hospital North West Ethiopia. Ethiop J Health Sci. 2009;19(3):173–177. [Google Scholar]
- 76.Bitew A, Admassie M, Getachew T. Spectrum and drug susceptibility profile of Bacteria recovered from patients with wound infection referred to Arsho advanced medical laboratory. Clin Med Res. 2018;7(1):8–17. doi: 10.11648/j.cmr.20180701.12 [DOI] [Google Scholar]
- 77.Hailu D, Derbie A, Mekonnen D, et al. Drug resistance patterns of bacterial isolates from infected wounds at Bahir Dar regional Health Research Laboratory center, Northwest Ethiopia. Ethiop J Health Devel. 2016;30(3):112–117. [Google Scholar]
- 78.Abraham Y, Wamisho BL. Microbial susceptibility of bacteria isolated from open fracture wounds presenting to the err of black-lion hospital, Addis Ababa University, Ethiopia. Afr J Microbiol Res. 2009;3(12):939–951. [Google Scholar]
- 79.Mulu A, Moges F, Tessema B, Kassu A. Pattern and multiple drug resistance of bacterial pathogens isolated from wound infection at University of Gondar Teaching Hospital, Northwest Ethiopia. Ethiop Med J. 2006;44(2):125–131. [PubMed] [Google Scholar]
- 80.Lema T, Woldeamanuel Y, Asrat D, et al. The pattern of bacterial isolates and drug sensitivities of infected ulcers in patients with leprosy in ALERT, Kuyera and Gambo hospitals, Ethiopia. Lepr Rev. 2012;83(1):40–51. doi: 10.47276/lr.83.1.40 [DOI] [PubMed] [Google Scholar]
- 81.Ayalew S. Bacterial profile and drug resistance pattern of pathogens isolated from wound infection at armed force referral and teaching Hospital, Addis Ababa, Ethiopia.2014. http://etd.aau.edu.et/handle/123456789/5552.
- 82.Mohammed A, Seid ME, Gebrecherkos T, Tiruneh M, Moges F. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. 2017;2017:1–10. doi: 10.1155/2017/8953829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Habte-Gabr E, Gedebou M, Kronvall G. Hospital-acquired infections among surgical patients in Tikur Anbessa hospital, Addis Ababa, Ethiopia. Am J Infect Control. 1988;16(1):7–13. doi: 10.1016/0196-6553(88)90004-1 [DOI] [PubMed] [Google Scholar]
- 84.Billoro BB, Nunemo MH, Gelan SE. Evaluation of antimicrobial prophylaxis use and rate of surgical site infection in surgical ward of Wachemo University Nigist Eleni Mohammed Memorial Hospital, Southern Ethiopia: prospective cohort study. BMC Infect Dis. 2019;19(298):1–8. doi: 10.1186/s12879-019-3895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guta M, Aragaw K, Merid Y. Bacteria from infected surgical wounds and their antimicrobial resistance in Hawassa University Referral Teaching Hospital, Southern Ethiopia. Afr J Microbiol Res. 2014;8(11):1118–1124. doi: 10.5897/AJMR2013.6544 [DOI] [Google Scholar]
- 86.Azene MK, Beyene BA. Bacteriology and antibiogram of pathogens from wound infections at Dessie Laboratory, North East Ethiopia. Tanzan J Health Res. 2011;13(4):68–74. doi: 10.4314/thrb.v13i4.64901 [DOI] [PubMed] [Google Scholar]
- 87.Sewunet T, Demissie Y, Mihret A, Abebe T. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiop J Health Sci. 2013;23(3):209–216. doi: 10.4314/ejhs.v23i3.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Motbainor H, Bereded F, Mulu W. Multi-drug resistance of blood stream, urinary tract and surgical site nosocomial infections of Acinetobacter baumannii and Pseudomonas aeruginosa among patients hospitalized at Felegehiwot referral hospital, Northwest Ethiopia: a cross-sectional study. BMC Infect Dis. 2020;20(92):1–11. doi: 10.1186/s12879-020-4811-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.CDC. Antibiotic Resistance Threats in the United States, 2019. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2019. [Google Scholar]
- 90.Martin E, Sanjay B, Bärbel C, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hygiene Infect Cont. 2017;12:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oliphant CM, Eroschenko K. Antibiotic resistance, Part 2: gram-negative pathogens. J Nurse Pract. 2015;11(1):79–86. doi: 10.1016/j.nurpra.2014.10.008 [DOI] [Google Scholar]
- 92.WHO. Antimicrobial resistance: global report on surveillance. World Health Organization 2014. Pp. 2
- 93.Ampaire L, Muhindo A, Orikiriza P, et al. A review of antimicrobial resistance in East Africa. Afr J Lab Med. 2016;5(1):1–6. doi: 10.4102/ajlm.v5i1.432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Breijyeh Z, Jubeh B, Karaman R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25(1340):1–23. doi: 10.3390/molecules25061340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.CDC. Tackling Drug-Resistant Infections Globally. Atlanta, GA: U.S. Department of Health and Human Services, CDC; 2016. [Google Scholar]
- 96.Le Doare K, Julia Bielicki PT. Heath, and Mike Sharland. Systematic Review of Antibiotic Resistance Rates Among Gram-Negative Bacteria in Children with Sepsis in Resource-Limited Countries J Ped Infec Dis Soc. 2015;4(1):11–20. [DOI] [PubMed] [Google Scholar]
- 97.BIOMérieux. Practical Guide to antimicrobial stewardship in hospitals. Available from https://www.biomerieux.co.uk/sites/subsidiary_uk/files/antimicrobial-stewardship-booklet-final.pdf. Accessed October17, 2020..
- 98.Brad S, John H, Eric P, Loren G, John E. Trends in antimicrobial drug development: implications for the future. Clinical Infectious Diseases. 2004;38(9):1279–1286. doi: 10.1086/420937 [DOI] [PubMed] [Google Scholar]
- 99.Nugent R, Back E, Beith A. A report of the centre for global development’s drug resistance working group. CGD. 2010. Pp. 27
- 100.Lim C, Takahashi E, Hongsuwan M, et al. Epidemiology and burden of multidrug resistant Bacterial infection in a developing country. eLife. 2016;5:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Brown PD. Multiple drug resistance in Staphylococcus aureus: a time-bomb for Jamaica and the region. Commonwealth Health Partnerships. 2014;74–77. [Google Scholar]
- 102.Jean C, Vincent J, Stephan H, Andreas V, Herman G, Pittet D. Ready for a world without antibiotics? The Pensières Antibiotic Resistance Call to Action. AMR Infec Cont. 2012;1(11):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Samuel SO, Kayode OO, Musa OI, et al. Nosocomial infections and the challenges of control in Developing countries. Afr J Cln Exper Microbiol. 11;2:102–110. [Google Scholar]