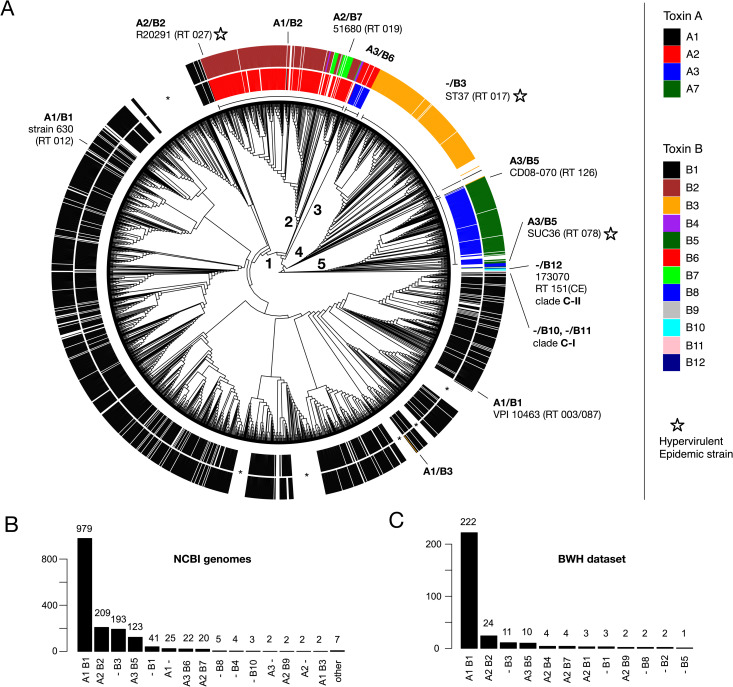
Fig 2. Toxin subtypes across the C. difficile phylogeny and occurrence of subtypes in a clinical CDI cohort.
(A) TcdA (inner ring) and TcdB (outer ring) subtypes mapped onto a tree of 1934 C. difficile genomes. The tree is a maximum likelihood phylogeny of NCBI-derived C. difficile genomes based on 14,194 genome-wide SNPs (see Methods). Lineages corresponding to previously identified C. difficile PaLoc clades (1–5) are labeled numerically. Selected clinically relevant strains are shown on the tree, with hypervirulent/epidemic outbreak strains indicated by stars. Asterisks indicate lineages without toxin genes. (B) Frequency of toxin subtypes detected in 1,934 representative, complete C. difficile genomes from NCBI/GenBank. A total of 1,640 (84.8%) C. difficile strains contained TcdA and/or TcdB, while 294 (15.2%) were toxin deficient. (c) Frequency of toxin subtypes detected in a CDI clinical cohort from Brigham and Women's Hospital (BWH). The total dataset contained 351 C. difficile genomes derived from infected patients. Of these, 289 (82.3%) contained toxin genes, and 62 (17.7%) were toxin deficient.