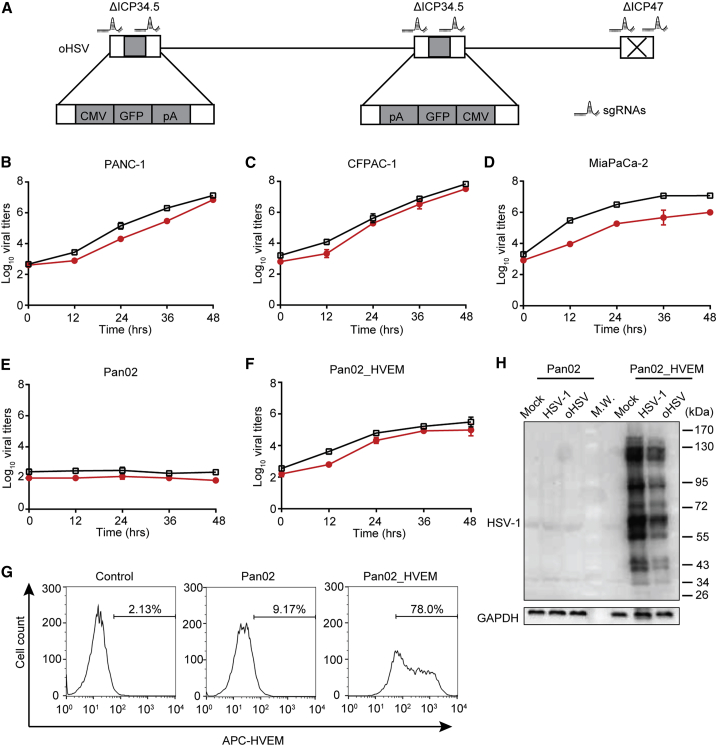
Figure 1.
Susceptibility of PDAC Cell Lines to Oncolytic Herpes Simplex Virus (oHSV) Infection
(A) Schematic diagram of the engineered oHSV genome. By CRISPR-Cas9-based homologous recombination with two sets of sgRNAs, two copies of the ICP34.5 gene were replaced by a GFP-expressing cassette, and ICP47 was deleted. (B–F)Different cell lines PANC-1 (B), CFPAC-1 (C), MiaPaCa-2 (D), Pan02 (E), and Pan02_HVEM (F) were infected with wild-type HSV-1 (black squares) or oncolytic HSV (oHSV) (red circles) at MOI = 0.01, and the replication of viruses in those cell lines was determined by titration assays at the indicated time points. Viral titers were presented as Log pfu per mL and plotted on the vertical axis. (G) Pan02 and Pan02_HVEM cells were stained with APC-conjugated anti-HVEM antibody and then subjected to flow cytometry analysis. The histograms were shown with cell count as vertical axis and fluorescence intensity of APC as horizontal axis. The percentage of HVEM-positive cells was labeled inside. Control stands for Pan02 cells stained with APC-conjugated isotype IgG. (H) Pan02 and Pan02_HVEM cells were mock infected or infected with indicated viruses for 24 h. The expression of viral proteins was analyzed by western blot with antibodies against total HSV-1 proteins (top panel) or GAPDH as an internal loading control (bottom panel), respectively. (B–F) Data represented the mean values from three independent experiments with standard deviation. (G and H) Data were representative for three independent experiments.