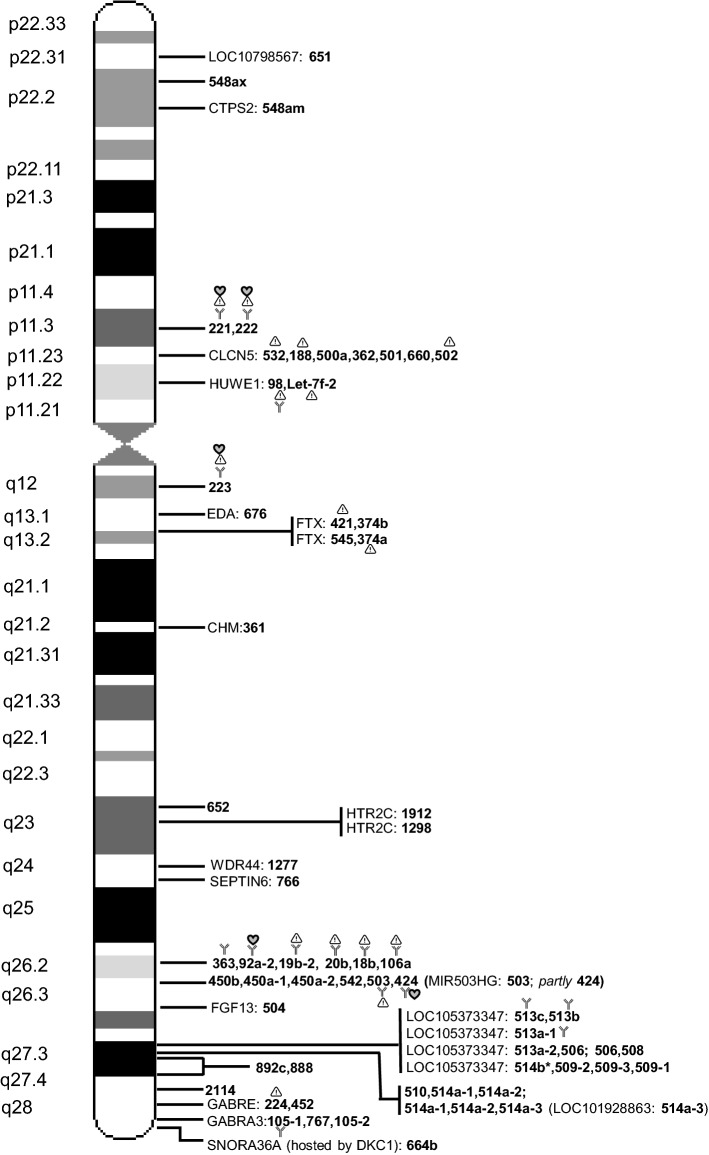
Fig. 1.
Map of microRNA sequences on human X chromosome. Information about validated miRNA sequence position is based on Ensembl and miRBase databases. On the left, chromosome bands are indicated and on the right, the miRNA names, simplified by eliminating « hsa-miR- » and indicated only by the number. Some miRNAs are intragenic and the host gene is indicated (name of host gene). Several miRNAs are clustered (within 10 kbp) and are shown on the same line separated by ‘,’, with the exception of miR-514b (*) that is located within 10.3 kbp of adjacent cluster; miRNA name is repeated when it can be considered belonging to different clusters. Some miRNAs are mainly involved in immunity ( ), cancer (
), cancer ( ) and cardiovascular homeostasis (
) and cardiovascular homeostasis ( ), as detailed in the text. LOC10798567, uncharacterized non-coding RNA; CTPS2, CTP synthase 2; CLCN5, chloride voltage-gated channel 5; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1; EDA, ectodysplasin A; FTX, FTX transcript, XIST regulator; CHM, CHM Rab escort protein; HTR2C, 5-hydroxytryptamine receptor 2C; WDR44, WD repeat domain 44; SEPTIN6, septin 6; MIR503HG, miR-503 host gene; FGF13, fibroblast growth factor 13; LOC105373347, periphilin-1-like; LOC101928863, uncharacterized non-coding RNA; GABRE, gamma-aminobutyric acid type A receptor epsilon subunit; GABRA3, gamma-aminobutyric acid type A receptor alpha3 subunit; SNORA36A, smal nucleolar RNA, H/ACA box 36A; DKC, dyskerin pseudouridine synthase 1.
), as detailed in the text. LOC10798567, uncharacterized non-coding RNA; CTPS2, CTP synthase 2; CLCN5, chloride voltage-gated channel 5; HUWE1, HECT, UBA and WWE domain containing E3 ubiquitin protein ligase 1; EDA, ectodysplasin A; FTX, FTX transcript, XIST regulator; CHM, CHM Rab escort protein; HTR2C, 5-hydroxytryptamine receptor 2C; WDR44, WD repeat domain 44; SEPTIN6, septin 6; MIR503HG, miR-503 host gene; FGF13, fibroblast growth factor 13; LOC105373347, periphilin-1-like; LOC101928863, uncharacterized non-coding RNA; GABRE, gamma-aminobutyric acid type A receptor epsilon subunit; GABRA3, gamma-aminobutyric acid type A receptor alpha3 subunit; SNORA36A, smal nucleolar RNA, H/ACA box 36A; DKC, dyskerin pseudouridine synthase 1.
The figure was inspired by Pinheiro et al. (2011) [35]