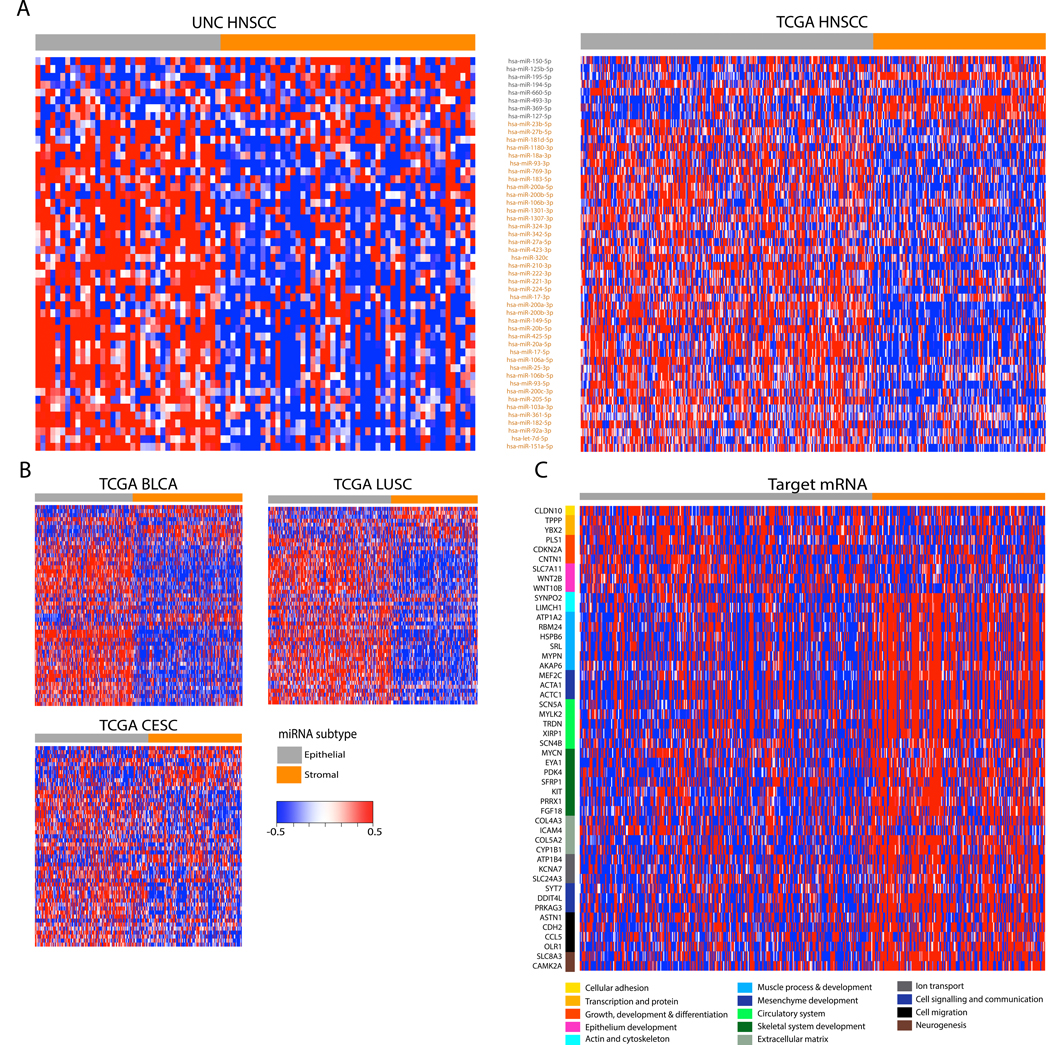
FIGURE 1:
MicroRNAs identify two HNSCC subtypes related to stages of epithelial differentiation. (A) Consensus clustering of 50 microRNAs with high integrative correlations between the 88 UNC samples (left) and the 474 TCGA samples (right) with HNSCC revealed two HNSCC subtypes (also see Supplementary Fig. 1). Columns represent individual samples and are labeled by class. Rows represent mature miRNA strands and are ordered by the discovery cohort hierarchical clustering. The epithelial-stromal developmental annotations of the two miRNA clusters are shown along with the miRNAs contained within each cluster. MicroRNAs downregulated in the ‘epithelial’ cluster are shown in gray, while those downregulated in the ‘stromal’ cluster are in orange. (B) Independent validation heatmaps show similar expression patterns of the two miRNA clusters and the two miRNA-based subtypes among the TCGA bladder urothelial carcinoma (BLCA), lung squamous cell carcinoma (LUSC), and cervical squamous cell carcinoma (CESC). Rows represent mature miRNA strands and are ordered the same as Fig. 1A. (C) The expression of curated, target predicted, and negatively correlated mRNAs are displayed as a heatmap along with biological function groupings. Columns have the same samples and order as the TCGA miRNA clustering in Fig. 1A.