Figure 4. Effect of myelomonocytic cells on cerebral repair and angiogenesis.
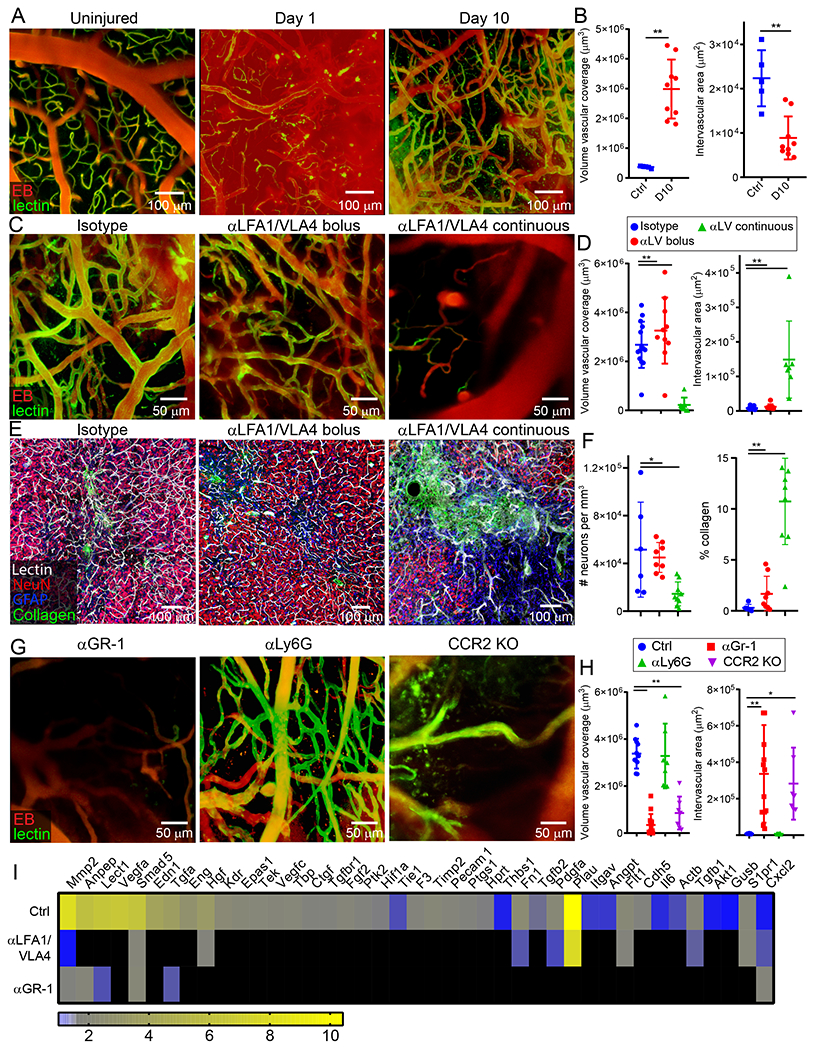
A-B. Intravital microscopy of cerebral vasculature and image-based quantification of vascular coverage and intervascular area shows development of new vessels with irregular distribution, increased diameter, and increased density at 10 d after injury relative to uninjured controls. Compilation from 2 independent experiments Ctrl n=5 and d10 n=9. C-D. A similar experiment was performed at 10 d post-injury following administration of continuous αLFA1/VLA4 (αLV), bolus αLFA1/VLA4, or isotype control antibodies. Compilation from 2 independent experiments Ctrl n=14, αLV n=10, 3 mice were not processed due to poor image quality. E-F. Confocal microscopy of brain sections from collagen-GFP mice 10 d after injury show extensive fibrosis (green) and NeuN+ neuronal (red) loss after continuous αLFA1/VLA4 administration relative to treatment with bolus αLFA1/VLA4 or isotype control antibodies. GFAP+ astrocytes are shown in blue. Compilation from 2 independent experiments Ctrl n=6, αLV n=8. G-H. Intravital microscopy of cerebral vasculature and image-based quantification of vascular coverage and intervascular area show lack of repair after continuous αGR-1 administration and in CCR2 KO mice but not after continuous αLy6G administration. Blood vessels were labeled with EB (red) and tomato-lectin (green). Compilation from 2 independent experiments Ctrl n=9, αGR-1 n=14, αLy6G, CCR2 KO n=7. In panels B, D, F, and H, graphs depict mean±SD (*P<0.05, **P<0.01, two-tailed Student’s t-test (B), One-way ANOVA/Tukey test (D,F,H)). I. Heatmap depicting qPCR analysis of genes encoding for angiogenesis-related proteins 6 d after injury shows downregulation of the angiogenesis pathway after continuous administration of αLFA1/VLA4 or αGr-1 relative to the isotype control group. Heatmap includes genes in which the difference between Ctrl and αLFA1/VLA4 had false discovery rate of Q<1% (multiple t-tests). Data are representative of 2 independent experiments with 4 mice per group per experiment. A comprehensive summary of the results and statistical analysis is provided in Extended Data Figure 9 and source data in Supplementary Table 4.
