Abstract
Alzheimer’s disease (AD) is characterized by the selective vulnerability of specific neuronal populations, the molecular signatures of which are largely unknown. To identify and characterize selectively vulnerable neuronal populations, we used single-nucleus RNA sequencing to profile the caudal entorhinal cortex and the superior frontal gyrus – brain regions where neurofibrillary inclusions and neuronal loss occur early and late in AD, respectively – from postmortem brains spanning the progression of AD-type tau neurofibrillary pathology. We identified RORB as a marker of selectively vulnerable excitatory neurons in the entorhinal cortex, and subsequently validated their depletion and selective susceptibility to neurofibrillary inclusions during disease progression using quantitative neuropathological methods. We also discovered an astrocyte subpopulation, likely representing reactive astrocytes, characterized by decreased expression of genes involved in homeostatic functions. Our characterization of selectively vulnerable neurons in AD paves the way for future mechanistic studies of selective vulnerability and potential therapeutic strategies for enhancing neuronal resilience.
INTRODUCTION
Selective vulnerability is a fundamental feature of neurodegenerative diseases, in which different neuronal populations show a gradient of susceptibility to degeneration. Selective vulnerability at the network level has been extensively explored in Alzheimer’s disease (AD)1–3. However, little is known about the mechanisms underlying selective vulnerability at the cellular level in AD, which could provide insight into disease mechanisms and lead to therapeutic strategies.
The entorhinal cortex (EC) is one of the first cortical brain regions to exhibit neuronal loss in AD4. Neurons in the external EC layers, especially in layer II, accumulate tau-positive neurofibrillary inclusions and die early in the course of AD5–10. However, these selectively vulnerable neurons have yet to be characterized at the molecular level. Furthermore, it is unknown whether there are differences in vulnerability among subpopulations of these neurons. Although rodent models of AD have offered important insights, the available models fail to simultaneously capture some critical disease processes, such as the accumulation of neurofibrillary inclusions and neuronal loss11, limiting the extrapolation of findings from rodent models to address selective vulnerability.
More recently, single-nucleus RNA-sequencing (snRNA-seq) has enabled large-scale characterization of transcriptomic profiles of individual cells from post-mortem human brain tissue12, 13. However, snRNA-seq studies of AD published to date have focused on cell-type specific differential gene expression between AD cases and healthy controls14, 15, without explicitly addressing selective vulnerability.
Here, we performed snRNA-seq on post-mortem brain tissue from a cohort of cases spanning the progression of AD-type tau neurofibrillary pathology to characterize changes in the relative abundance of cell types and cell type subpopulations. We discovered a selectively vulnerable subpopulation of excitatory neurons in the EC and validated the selective depletion of this subpopulation during AD progression with quantitative histopathology, using multiplex immunofluorescence in EC regions delineated by rigorous cytoarchitectonic criteria. Furthermore, we uncovered an astrocyte subpopulation likely corresponding to reactive astrocytes that showed downregulation of genes involved in homeostatic function.
RESULTS
Cohort selection and cross-sample alignment
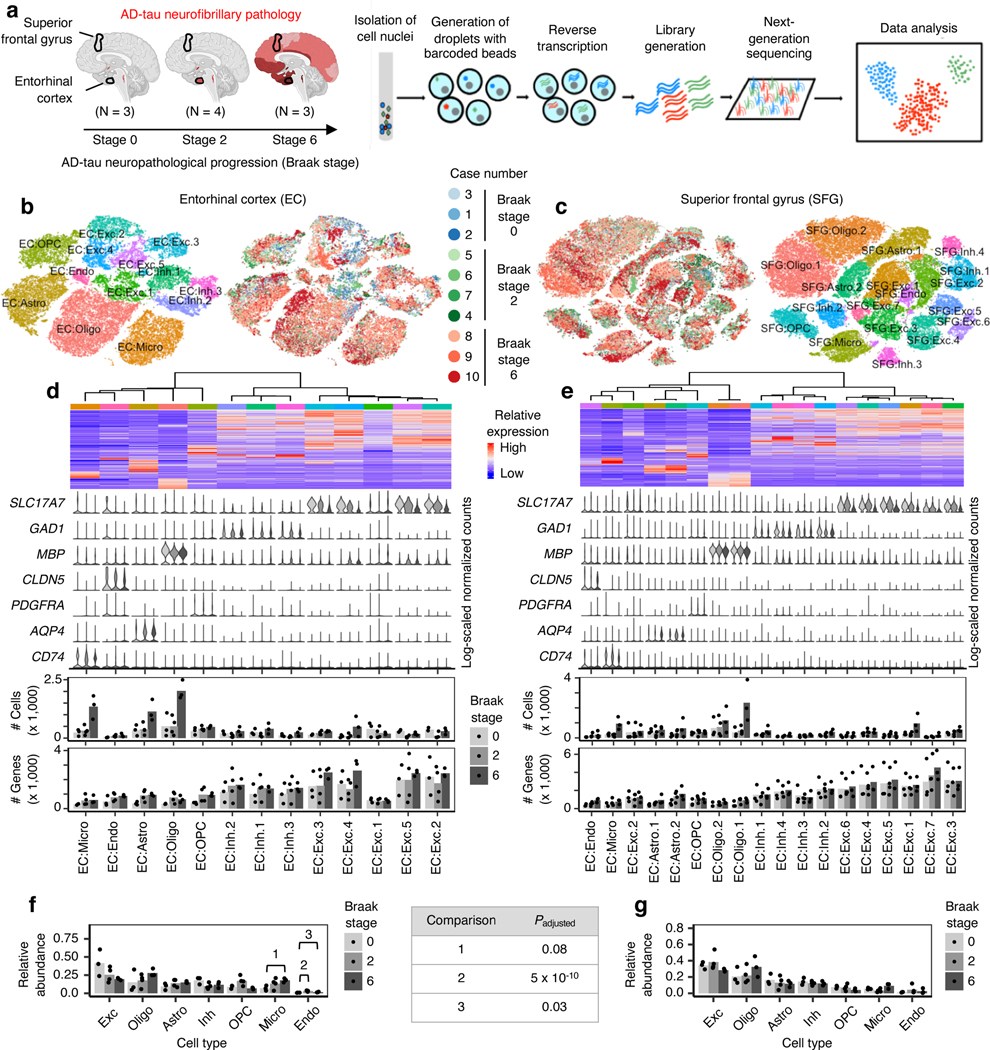
We performed snRNA-seq on cell nuclei extracted from postmortem brain tissue (see Methods) from the EC at the level of the mid-uncus and from the superior frontal gyrus (SFG) at the level of the anterior commissure (Brodmann area 8), from 10 male APOE ε3/ε3 individuals representing the cortical-free, early and late stages of AD-type tau neurofibrillary pathology (Braak stages1 0, 2 and 6; Fig. 1a, Table 1). The neuropathological hallmarks of AD are amyloid plaquesand neurofibrillary inclusions. Since the accumulation of neurofibrillary inclusions measured by the Braak staging system is the best correlate of clinical cognitive decline, after neuronal loss16, we reasoned that profiling matched EC and SFG samples across different Braak stages would allow us to isolate the effect of disease progression on cell types and cell type subpopulations.
Fig. 1 |. AD progression differentially affects the cell-type composition of the EC and SFG.
a, Schematic of experimental design and sample processing. Darker shades of red in brain cartoons reflect more severe AD-tau neurofibrillary pathology. b-c, tSNE projection of cells from the EC (b) and SFG (c) in their respective alignment spaces, colored by individual of origin (center) or cluster assignment (outer). d-e, Heatmap and hierarchical clustering of clusters and cluster marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Expression of cell type markers in each cluster (second subpanel). The average number of cells and average number of genes detected per cell in each cluster (third and fourth subpanels). f-g, Relative abundance of major cell types across Braak stages. For each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Cell type abbreviations: Exc – excitatory neurons, Oligo – oligodendrocytes, Astro – astrocytes, Inh – inhibitory neurons, OPC – oligodendrocyte precursor cells, Micro – microglia, Endo – endothelial cells.
Table 1 |.
Description of post-mortem cohort.
| Cases used for snRNA-seq | ||||||||
|---|---|---|---|---|---|---|---|---|
| Case # | Braak stage | Sex | Age at death (years) | Post-mortem interval (hours) | ADNC score | CDR before death | APOE genotype | Source |
| 1 | 0 | M | 50 | 13 | A0,B0,C0 | 0 | E3/E3 | BBAS |
| 2 | 0 | M | 60 | 12 | A0,B0,C0 | 0.5 | E3/E3 | BBAS |
| 3 | 0 | M | 71 | 12 | A1,B0,C0 | 0 | E3/E3 | BBAS |
| 4 | 2 | M | 72 | 15 | A1,B1,C0 | 0 | E3/E3 | BBAS |
| 5* | 2 | M | 77 | 4.9 | A2,B1,C1 | 0.5 | E3/E3 | UCSF |
| 6* | 2 | M | 87 | 30 | A2,B1,C2 | 2 | E3/E3 | UCSF |
| 7* | 2 | M | 91 | 50 | A1,B1,C1 | 0 | E3/E3 | UCSF |
| 8* | 6 | M | 72 | 6.9 | A3,B3,C3 | 3 | E3/E3 | UCSF |
| 9* | 6 | M | 82 | 6.7 | A3,B3,C3 | 3 | E3/E3 | UCSF |
| 10 | 6 | M | 82 | 9 | A3,B3,C3 | 3 | E3/E3 | UCSF |
| Cases used for immunofluorescence validation | ||||||||
| Case # | Braak stage | Sex | Age at death | Post-mortem interval (hours) | ADNC score | CDR before death | APOE genotype | Source |
| 5* | 2 | M | 77 | 4.9 | A2,B1,C1 | 0.5 | E3/E3 | UCSF |
| 6* | 2 | M | 87 | 30 | A2,B1,C2 | 2 | E3/E3 | UCSF |
| 7* | 2 | M | 91 | 50 | A1,B1,C1 | 0 | E3/E3 | UCSF |
| 8* | 6 | M | 72 | 6.9 | A3,B3,C3 | 3 | E3/E3 | UCSF |
| 9* | 6 | M | 82 | 6.7 | A3,B3,C3 | 3 | E3/E3 | UCSF |
| 11 | 0 | F | 62 | 10.1 | A1,B0,C0 | 0 | NA | BBAS |
| 12 | 0 | M | 64 | 12 | A0,B0,C0 | 0 | E3/E3 | BBAS |
| 13 | 1 | M | 60 | 19 | A0,B1,C0 | 0 | NA | BBAS |
| 14 | 1 | F | 64 | 13 | A1,B1,C0 | 0 | E3/E3 | BBAS |
| 15 | 1 | M | 70 | 11 | A1,B1,C0 | 0 | E3/E3 | BBAS |
| 16 | 1 | F | 82 | 9.6 | A1,B1,C0 | 0 | NA | BBAS |
| 17 | 2 | F | 79 | 18 | A1,B1,C1 | 0 | E3/E3 | BBAS |
| 18 | 2 | F | 81 | 30.3 | A1,B1,C0 | NA | E3/E3 | UCSF |
| 19 | 3 | M | 81 | 8.3 | A2,B2,C3 | 1 | NA | UCSF |
| 20 | 3 | M | 84 | 28 | A3,B2,C2 | 1 | NA | UCSF |
| 21 | 3 | F | 88 | 9.8 | A3,B2,C2 | 0.5 | E3/E3 | UCSF |
| 22 | 3 | M | 89 | 9.1 | A3,B2,C2 | 1 | E3/E3 | UCSF |
| 23 | 4 | F | 87 | 9.5 | A1,B2,C3 | 2 | E3/E3 | UCSF |
| 24 | 4 | M | 91 | 11.2 | A3,B2,C2 | 0.5 | E3/E3 | UCSF |
| 25 | 4 | M | 103 | 7.8 | A1,B2,C2 | NA | E3/E3 | UCSF |
| 26 | 5 | M | 77 | 8.4 | A3,B3,C3 | 0.5 | E4/E4 | UCSF |
| 27 | 5 | M | 85 | 11.2 | A3,B3,C3 | 1 | E3/E3 | UCSF |
| 28 | 5 | M | 86 | 8.6 | A3,B3,C3 | 2 | E3/E4 | UCSF |
| 29 | 5 | F | 87 | 17 | A3,B3,C2 | 3 | E3/E3 | BBAS |
| 30 | 6 | F | 64 | 7.3 | A3,B3,C3 | 3 | E3/E4 | UCSF |
| 31 | 6 | F | 67 | 9.7 | A3,B3,C3 | 3 | E4/E4 | UCSF |
Asterisks denote cases used both for snRNA-seq and immunofluorescence validation. The AD neuropathological change (ADNC) score incorporates assessment of amyloid-beta deposits (“A”), staging of neurofibrillary tangles (“B”), and scoring of neuritic plaques (“C”)48. The Clinical Dementia Rating (CDR) reflects the degree of cognitive impairment49.
A challenge in characterizing the impact of disease progression on different cell type subpopulations is that these subpopulations need to be defined in a way that is independent from the effect of disease progression. Typically, cell type subpopulations are defined by sub-grouping cells of the same cell type through cluster analysis (i.e. clustering), followed by examination of marker gene expression in the resulting clusters. To remove the effect of disease progression on clustering, we performed, prior to clustering, cross-sample alignment of the data from each brain region using scAlign (see Methods). Importantly, after identifying clusters in the alignment space, we used the original data for subsequent analyses involving examination of gene expression, such as identifying differentially expressed genes between clusters.
Changes in broad cell type composition with neuropathological AD progression
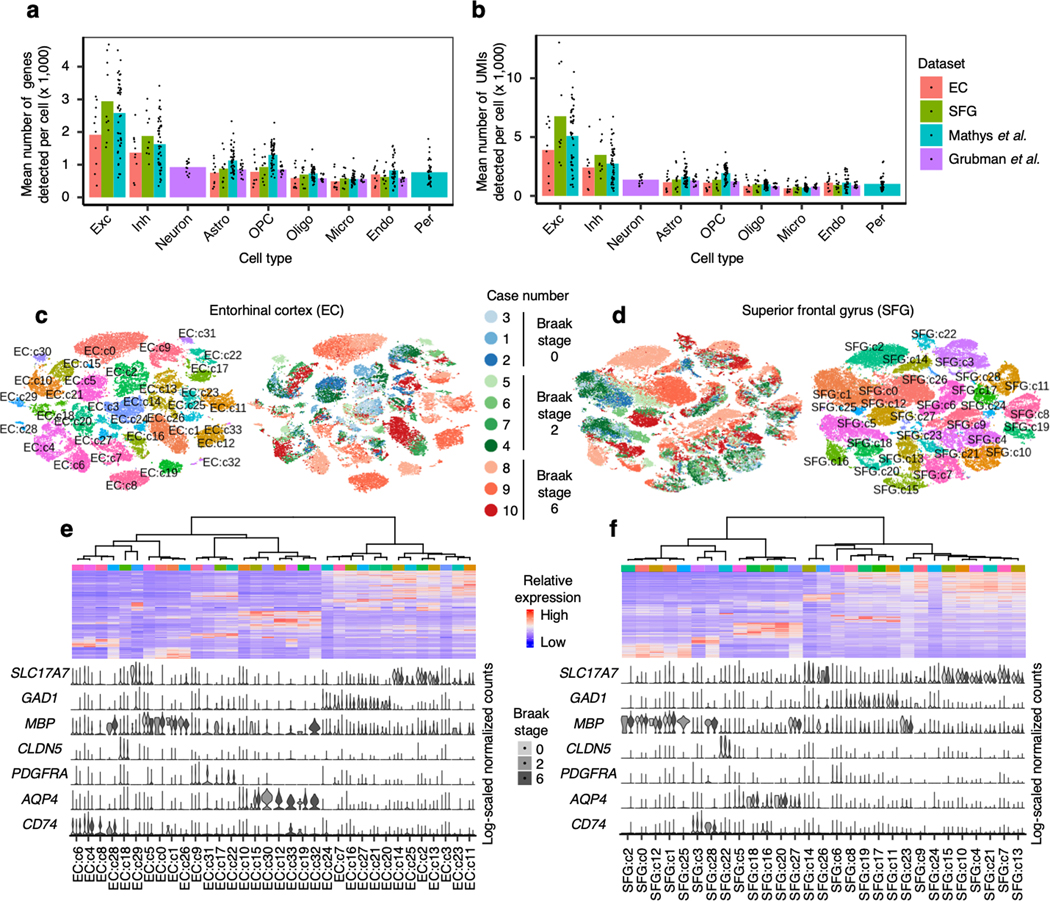
After quality control (see Methods), we recovered 42,528 cells from the EC and 63,608 cells from the SFG. Examination of the average number of genes and unique molecular identifiers (UMIs) detected per cell showed similar or superior transcript coverage compared to previously published AD snRNA-seq datasets14, 15 (Extended Data Fig. 1a,b).
After cross-sample alignment, we performed clustering and recovered clusters that demonstrated spatial grouping in t-stochastic neighborhood embedding (tSNE) largely uncorrelated with the individual of origin (Fig. 1b,c). Furthermore, the clusters showed specific expression of cell type markers and grouped in a manner consistent with their expression of cell type markers in hierarchical clustering (Fig. 1d,e, see Methods). For comparison, we also performed clustering without cross-sample alignment, which resulted in many clusters that were defined by individual of origin in addition to cell type (Extended Data Fig. 1c–f). Having confirmed the effectiveness of cross-sample alignment in removing the effect of technical and experimental factors on clustering, we then assigned clusters to broad cell types (i.e. excitatory neurons, inhibitory neurons, astrocytes, oligodendrocytes, oligodendrocyte precursor cells, microglia, and endothelial cells) based on their expression of cell type markers (Fig. 1d,e, see Methods).
Next, to assess whether the proportions of broad cell types change with disease progression, we aggregated clusters assigned to the same cell type for each individual and then computed the relative abundance of each cell type in each individual. We tested for statistical significance using beta regression and corrected for multiple testing using Holm’s method (see Methods). While there were not many statistically significant changes in the relative abundance of cell types, we observed a downward trend in the relative abundance of EC excitatory neurons in Braak stages 2 (Punadjusted = 0.18) and 6 (Punadjusted = 0.02), and of SFG excitatory neurons only in Braak stage 6 (Punadjusted = 0.05), consistent with early involvement of the EC and sparing of the SFG until late Braak stages, and the previously described greater vulnerability of excitatory neurons relative to inhibitory neurons in AD17, 18.
Selective vulnerability of excitatory neuron subpopulations
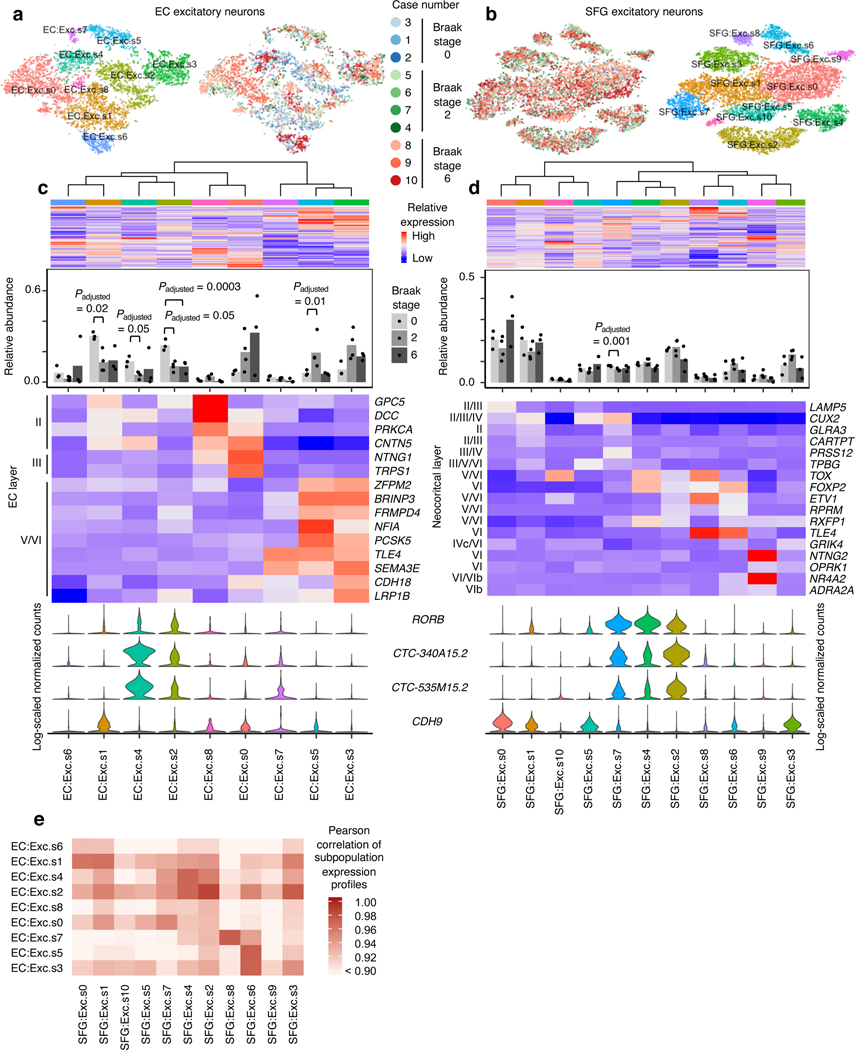
Based on these observations, we next asked whether specific subpopulations of excitatory neuron show a decline in their relative abundance with disease progression, by performing subclustering of excitatory neurons in the EC and SFG after cross-sample alignment (see Methods). The EC, a relatively phylogenetically conserved brain structure in mammals, is among the first cortical fields to accumulate tau-positive neurofibrillary inclusions followed by neuronal loss in AD1. The EC is a heterogeneous structure and cytoarchitectonic considerations matter when analyzing and sampling this region to avoid biased observations19. During evolution, the position of the EC changed, and the mouse medial EC (the source of our layer-specific marker genes) is generally regarded as the equivalent of the caudal EC in humans (our sampling location)20, 21.
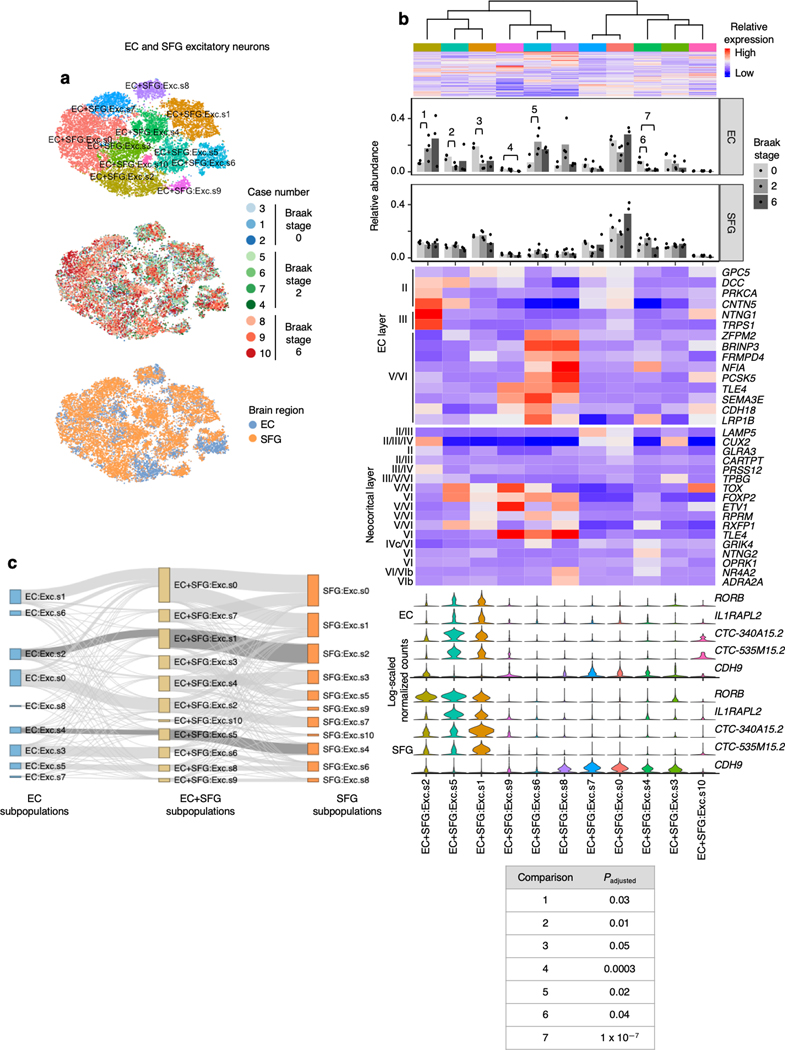
In the EC, we discerned nine excitatory neuron subpopulations (Fig. 2a–d). These subpopulations exhibited distinct expression of EC layer-specific genes identified in the mouse medial EC22. Notably, subpopulation EC:Exc.s2 showed a striking ~50% decrease in its relative abundance in Braak stage 2 compared to Braak stage 0, with no further decrease in Braak stage 6 (Fig. 2c), suggesting depletion early in disease. EC:Exc.s1 and EC:Exc.s4 similarly exhibited a ~50–60% reduction in their relative abundance in Braak stage 2. EC:Exc.s1, EC:Exc.s2, and EC:Exc.s4 expressed genes associated with mouse EC layer II (Fig. 2c), consistent with the fact that tau neurofibrillary inclusions are known to accumulate preferentially in human EC layer II early in AD5–8. However, not all subpopulations expressing genes associated with mouse EC layer II showed similar levels of early vulnerability. For example, EC:Exc.s6 and EC:Exc.s8 did not demonstrate statistically significant changes in their relative abundance across disease progression. We failed to find evidence of selective vulnerability in neuronal subpopulations expressing genes associated with mouse EC layer III (EC:Exc.s0) or V/VI (EC:Exc.7, EC:Exc.s5, EC:Exc.s3). In fact, EC:Exc.s5 exhibited a statistically significant increase in its relative abundance in Braak stage 2. Since neurons are post-mitotic, this increase is likely due to the selective earlier depletion of more vulnerable excitatory neuron subpopulations, followed by later depletion of EC:Exc.s5.
Fig. 2 |. RORB-expressing excitatory neuron subpopulations in the EC are selectively vulnerable.
a-b, tSNE projection of excitatory neurons from the EC (a) and SFG (b) in their respective alignment spaces, colored by individual of origin (center) or subpopulation identity (outer). c-d, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations across Braak stages (second subpanel); for each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of EC layer-specific genes identified from Ramsden et al.22 (c, third subpanel). Expression heatmap of neocortical layer-specific genes from Lake et al.12 (d, third subpanel). Expression of selectively vulnerable subpopulation markers identified in the EC (bottom subpanel). e, Heatmap of Pearson correlation between the gene expression profiles of EC and SFG subpopulations.
To identify molecular markers of selectively vulnerable excitatory neuron subpopulations in the EC (EC:Exc.s2, EC:Exc.s4, EC:Exc.s1), we inspected transcript levels of genes differentially expressed between pairs of subpopulations and curated a set of genes which were specifically expressed by no more than four subpopulations (Extended Data Fig. 2a), which we decided was a reasonable threshold for a positive marker to be useful. We found that EC:Exc.s2 and EC:Exc.s4 specifically expressed RORB, CTC-340A15.2 and CTC-535M15.2 (Fig. 2c). RORB (RAR-related Orphan Receptor B) encodes a transcription factor known as a marker and developmental driver of layer IV neurons in the neocortex23–25, but is also expressed by neurons in other layers13. Little is known about the non-coding transcripts CTC-340A15.2 and CTC-535M15.2 in the context of neuronal identity and function. We also found that EC:Exc.s1 was marked by high expression of CDH9 (Fig. 2c), which encodes a cadherin with neuron-specific expression. However, CDH9 was also expressed by other excitatory neuron subpopulations in the EC, and we could not find markers that were specifically expressed only in EC:Exc.s1. Therefore, we chose to focus our analysis on EC:Exc.s2 and EC:Exc.s4.
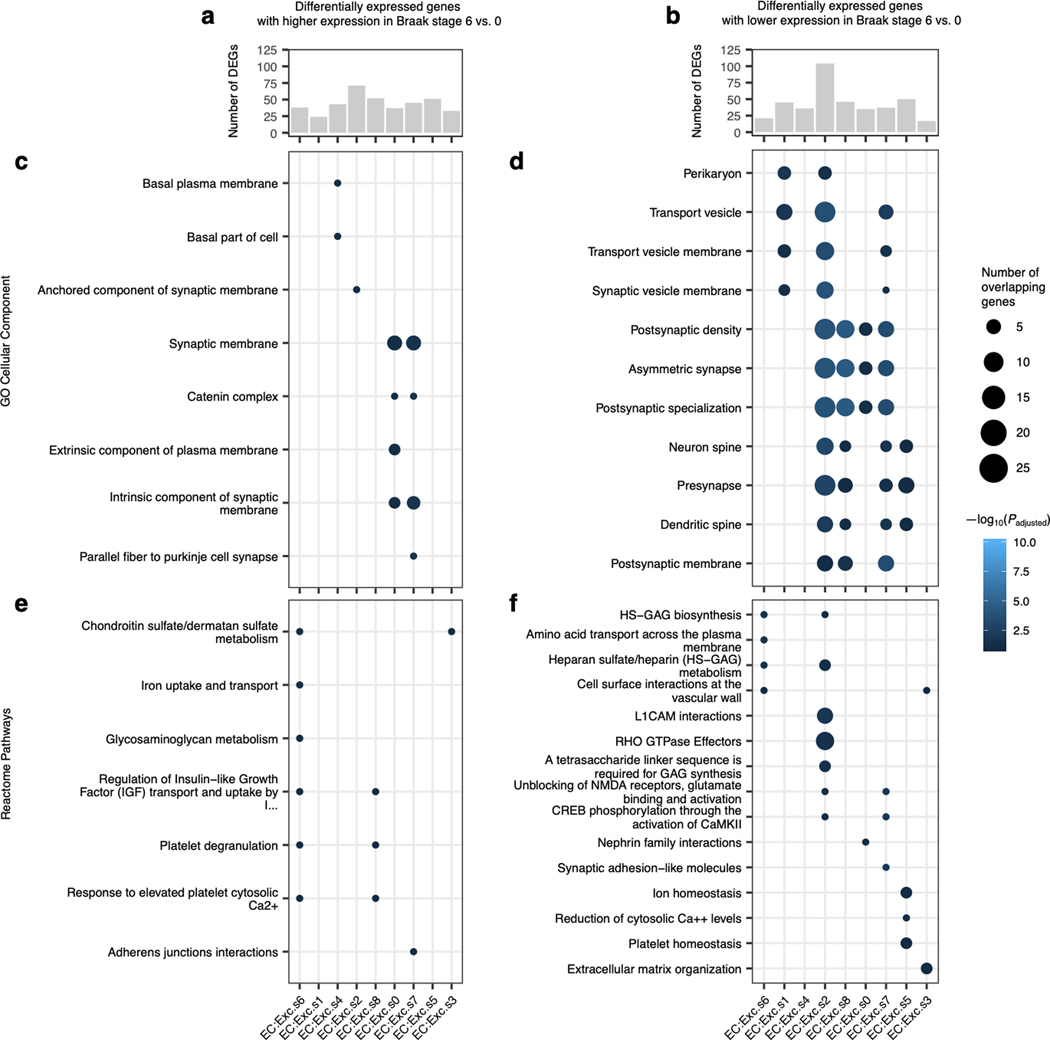
In addition to identifying molecular markers of the selectively vulnerable EC:Exc.s2 and EC:Exc.s4 neurons, we also enumerated genes that were differentially expressed in EC:Exc.s2 and EC:Exc.s4 compared to all other excitatory neurons in the EC, controlling for differences across individuals (see Methods). We found that genes with higher expression in EC:Exc.s2 and EC:Exc.s4 were enriched for axon-localized proteins and voltage-gated potassium channels, whereas genes with lower expression in EC:Exc.s2 and EC:Exc.s4 were enriched for synapse- and dendrite-localized proteins and pathways involving G-protein mediated signaling, ion transport, and neurotransmitter receptor signaling (Extended Data Fig. 2b–e, Supplementary Table 1).
We also performed differential gene expression analysis across Braak stages for EC excitatory neuron subpopulations (see Methods), comparing Braak stage 6 vs. 0, which yielded the largest number of differentially expressed genes. We found a broad decrease in expression of genes encoding pre- and post-synaptic proteins in Braak stage 6 vs. 0 for many EC excitatory neuron subpopulations (Extended Data Fig. 3b,d,f; Supplementary Table 2). Furthermore, we observed that the selectively vulnerable subpopulation EC:Exc.s2 had the largest number of downwardly differentially expressed genes and the strongest enrichments for pre- and post-synaptic proteins in these genes (Extended Data Fig. 3b,d). Overall, the downregulation of synapse-related genes we have observed mirrors the findings from a recent preprint by Marinaro et al.26, which examined the frontal cortex in familial monogenic AD using snRNA-seq, and is consistent with a previous study of gene expression changes in AD in the entorhinal cortex and other brain regions employing laser capture microdissection of neurons followed by DNA microarray analysis27.
We next examined excitatory neuron subpopulations in the SFG. Similar to previous studies12, 13, we found that excitatory neuron subpopulations in the SFG (11 in total) expressed distinct sets of neocortical layer-specific genes (Fig. 2b,d), recapitulating the laminar organization of the neocortex. Interestingly, SFG:Exc.s4 and SFG:Exc.s2, which were marked by the vulnerability markers we identified in the EC (RORB, CTC-340A15.2 and CTC-535M15.2), trended towards decreased relative abundance only in Braak stage 6 (Fig. 2d), consistent with the late appearance of neurofibrillary inclusions in the SFG starting at Braak stage 5. Although SFG:Exc.s7, which also expressed the EC vulnerability markers, exhibited a statistically significant decrease in relative abundance in Braak stage 2 but not 6, the magnitude of change was negligibly small.
Given that SFG:Exc.s4 and SFG:Exc.s2 expressed similar markers as EC:Exc.s4 and EC:Exc.s2, we wondered if SFG:Exc.s4 and SFG:Exc.s2 may resemble EC:Exc.s4 and EC:Exc.s2 more broadly at the transcriptome level. SFG:Exc.s4 and SFG:Exc.s2 were indeed most similar to EC:Exc.s4 and EC:Exc.s2 based on the Pearson correlation coefficient between the expression profiles of SFG and EC subpopulations (Fig. 2e). We observed the same pattern when we mapped subpopulations in the EC to those in the SFG by performing cross-sample alignment for both brain regions jointly (Extended Data Fig. 4). This similarity is consistent with the reported similarity between deep layer neocortical excitatory neurons and EC excitatory neurons in general28. The similarity in transcriptomes of vulnerable excitatory neurons in different brain regions is intriguing and suggests similar mechanisms of selective vulnerability in different brain regions.
Although the decrease in the relative abundance of SFG:Exc.s2 and SFG:Exc.s4 in Braak stage 6 was not statistically significant after correction for multiple testing, we asked if we could detect signs of selective vulnerability in neocortical RORB-expressing excitatory neurons in an independent dataset with a larger sample size. To this end, we reanalyzed data from Mathys et al.14, which profiled the prefrontal cortex from 24 AD cases and 24 healthy controls, with our cross-sample alignment pipeline and performed subclustering of excitatory neurons. In the Mathys et al. dataset14, we discerned 10 excitatory neuron subpopulations, each of which expressed distinct sets of neocortical layer-specific genes (Extended Data Fig. 5a,b) similar to Lake et al.12 and our dataset. Of these 10 subpopulations, Mathys:Exc.s4, Mathys:Exc.s5, and Mathys:Exc.s1 expressed RORB at high levels (CTC-340A15.2 and CTC-535M15.2 were not available in the pre-processed Mathys et al.14 data). Importantly, we observed a statistically significant decrease in the relative abundance of Mathys:Exc.s4 in male AD cases vs. controls (Extended Data Fig. 5b), recapitulating the selective vulnerability observed in our dataset, which consists only of male individuals. Furthermore, gene expression correlation analysis showed that Mathys:Exc.s4 was the most similar to EC:Exc.s2 and EC:Exc.s4 (Extended Data Fig. 5c), again demonstrating similarity between selectively vulnerable excitatory neurons in the neocortex and those in the EC.
Although we did not detect any statistically significant changes in the relative abundance of RORB-expressing subpopulations in female individuals in Mathys et al.14, Mathys.Exc.s1 trended towards decreased relative abundance in female AD cases (Punadjusted = 0.17) and mapped to EC:Exc.s2 by gene expression correlation (Extended Data Fig. 5b,c). Furthermore, Marinaro et al.26 included both male and female cases of monogenic AD and also reported the selective vulnerability of two out of four RORB-expressing excitatory neuron subpopulations in the prefrontal cortex (ExcB1 and ExcB4)26, providing further evidence that subsets of RORB-expressing excitatory neurons in the neocortex are selectively vulnerable.
Considering the Mathys et al.14 and the Marinaro et al.26 datasets together with our dataset, it appears that while not all RORB-expressing excitatory neuron subpopulations in the neocortex showed signs of selective vulnerability, those that did were the most similar to RORB-expressing excitatory neurons in the EC, all of which showed signs of selective vulnerability.
Validation of the selective vulnerability of RORB-expressing excitatory neurons
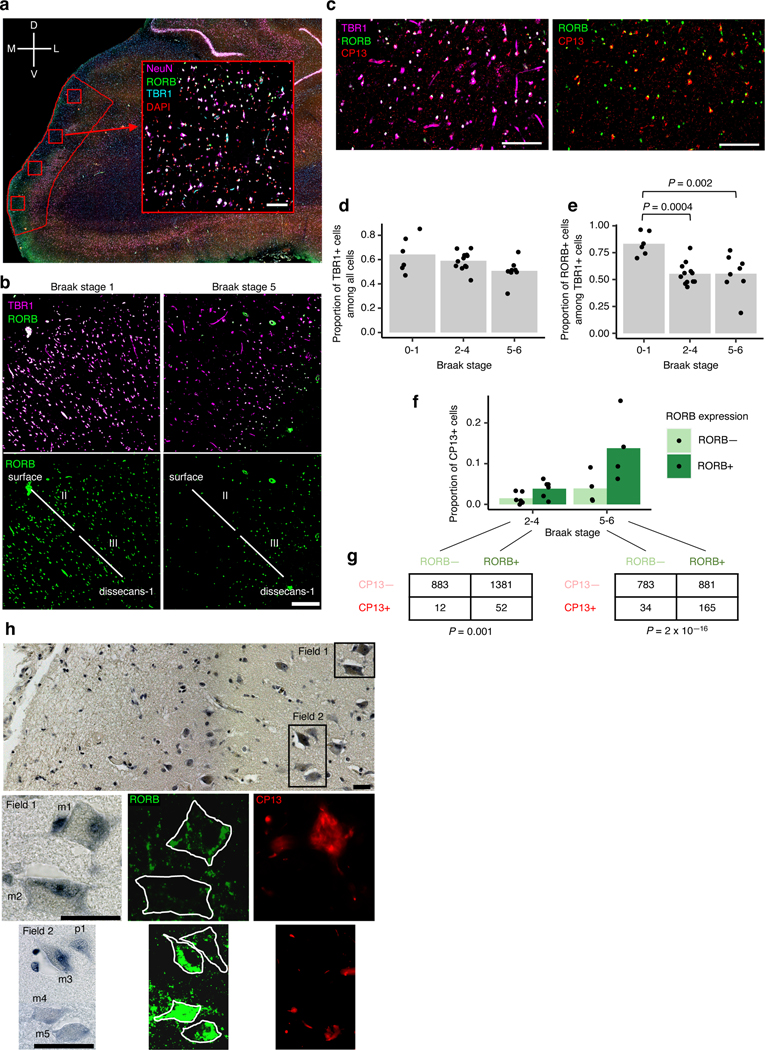
To validate our finding that RORB-expressing excitatory neurons in the EC were highly vulnerable in AD, we performed multiplex immunofluorescence on postmortem brains of 26 individuals spanning Braak stages 0 to 6, who were devoid of non-AD neuropathological changes (Table 1). Specifically, we quantified the proportion of excitatory neurons and RORB-positive excitatory neurons in the EC superficial layers (i.e. above layer IV, which we also refer to as dissecans-119 in Fig. 3b). Given EC heterogeneity, we used rigorous cytoarchitectonic parameters to delineate the caudal EC and minimize artifactual results (Fig. 3a–c, Extended Data Fig. 6, see Methods). We used multiplex immunofluorescence29 to label nuclei (DAPI), excitatory neurons (TBR1), RORB+ neurons, and phospho-tau neuronal inclusions (CP-13, Ser 202). We observed a substantial reduction in the proportion of RORB+ neurons among excitatory neurons in Braak stages 2–4 and 5–6 compared to Braak stages 0–1 (Fig. 3d,e). Furthermore, by analyzing a subset of cases, we detected phospho-tau (CP-13) preferentially in RORB+ compared to RORB- excitatory neurons (Fig. 3f–g). Thus, our results substantiate that RORB-expressing excitatory neurons are highly vulnerable in AD and support a model in which their depletion is a consequence of accumulating tau neurofibrillary changes.
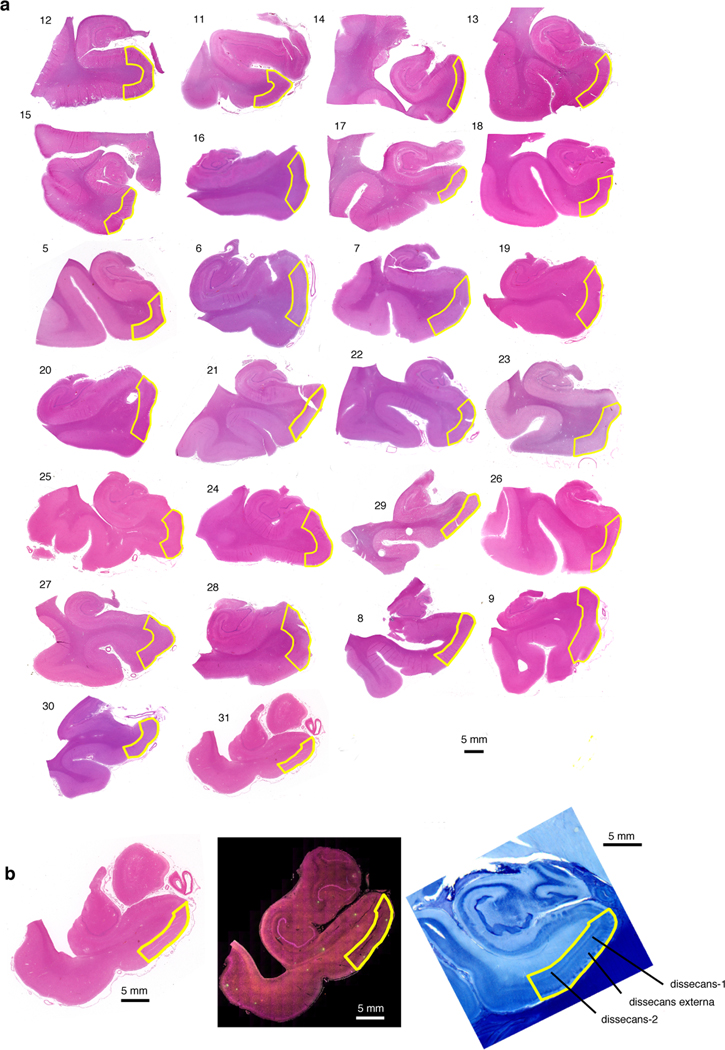
Fig. 3 |. Immunofluorescence of the EC validates selective vulnerability of RORB-expressing excitatory neurons.
a, The method for extracting regions of interest (ROI) is illustrated using a representative brain slice used for immunofluorescence (pseudo-colored: DAPI in blue, RORB in green, TBR1 In orange and NeuN in pink) with the EC delineated in red. Four ROIs (drawn in red squares) were randomly distributed along the superficial layers of the EC and extracted for quantification after masking neurons (see Methods). A representative ROI image is shown as insert (note that the pseudo-coloring scheme for the insert, as indicated in the Figure, differs from the pseudo-coloring scheme of the larger panel). The anatomical orientation of the slice is provided in the top left corner (D – dorsal, V – ventral, M – medial, L – lateral). b, Representative RORB staining in a Braak stage 1 sample (left) vs. a Braak stage 5 sample (right), shown with (top) and without (bottom) excitatory neurons marked by TBR1 staining. The EC layers captured in the image are demarcated in the bottom subpanels (see Methods and Extended Data Fig. 6). c, Representative CP13 staining in a Braak stage 6 sample, shown together with TBR1 and RORB staining (left) or only with RORB staining (right). d-e, Proportion of TBR1+ cells among all cells (d) or proportion of RORB+ cells among TBR1+ cells (e) averaged across ROIs for each individual across groups of Braak stages; statistical significance of differences in the above proportions across groups of Braak stages (Braak 0–1 n=6, Braak 2–4 n=12, Braak 5–6 n=8, where n is the number of individuals sampled) was determined by beta regression without adjustment for multiple comparisons. f, Proportion of CP13+ cells in RORB- or RORB+ excitatory neurons (i.e. TBR1+ cells) averaged across ROIs for each individual across groups of Braak stages. g, Contingency tables of raw counts of TBR1+ cells based on their RORB or CP13 staining status summed across ROIs and individuals for each group of Braak stages (Braak 2–4 n=6, Braak 5–6 n=4, where n is the number of individuals sampled); the Fisher’s Exact Test p-value (two-sided) is shown below each table. h, Representative image of EC layer II neurons stained with gallocyanin (top subpanel) with the corresponding RORB and CP13 immunofluorescence signal shown in selected fields (Field 1 – middle subpanels, Field 2 – bottom subpanels). RORB+ neurons include both large multipolar neurons (m1, m3, m4, m5) and pyramidal neurons (p1). One large multipolar neuron (m2) is RORB-. The neuronal somas are outlined manually in white in the RORB immunofluorescence images to aid interpretation. Scale bars shown in a-c correspond to 100 microns; scale bars shown in h correspond to 15 microns. For all data shown in this figure, the experiment was performed once.
Given that largel multipolar neurons of “stellate” morphology in EC layer II are particularly vulnerable in AD5–8, we examined the morphological features of layer II’s RORB+ excitatory by overlaying immunofluorescence with Nissl staining. We found that RORB+ excitatory neurons adopted various shapes, including pyramidal and multipolar morphologies (Fig. 3h). Conversely, some large multipolar neurons are RORB-negative (Fig. 3h). Our results are consistent with the known vulnerability of large multipolar EC layer II neurons and demonstrate that molecular characterization of vulnerable neurons refines the results of morphological studies.
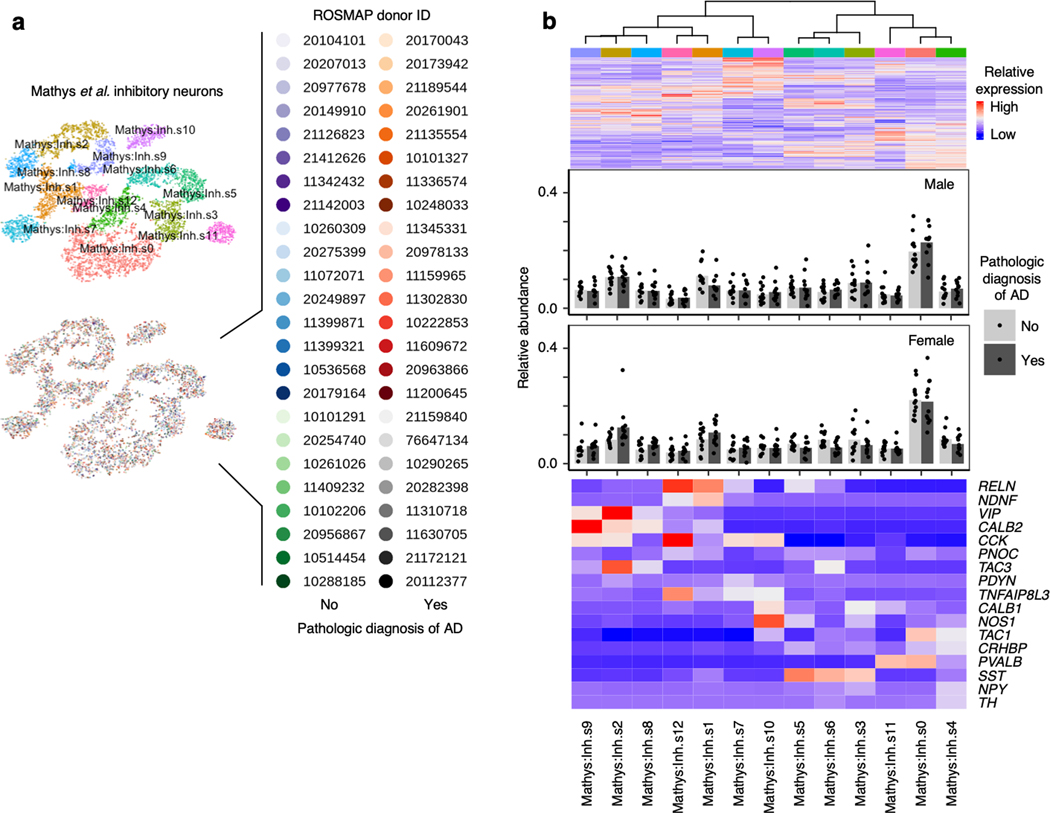
Lack of differences in vulnerability of inhibitory neuron subpopulations
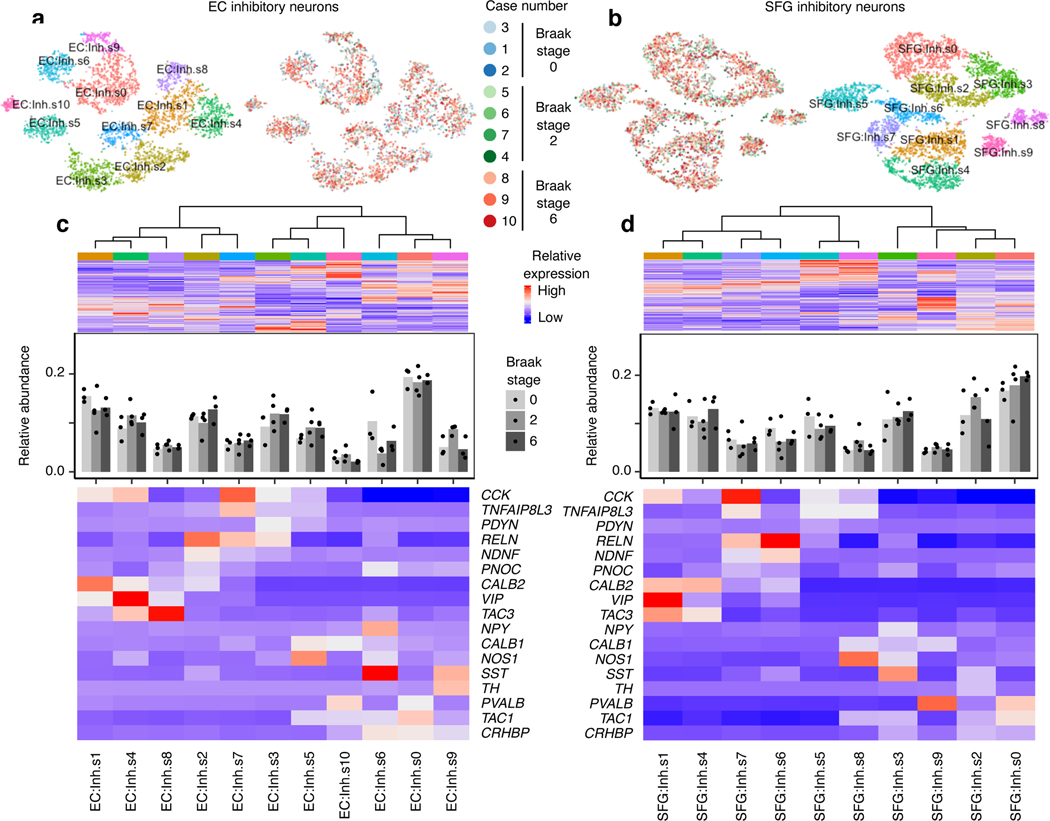
We next examined inhibitory neurons, which are more resistant to tau pathology than excitatory neurons in AD17, 18. In both brain regions, inhibitory neuron subpopulations expressed distinct sets of inhibitory neuron subtype markers (Fig. 4a–d), consistent with previous studies12, 13. We did not any detect statistically significant changes in the relative abundance of inhibitory neurons subpopulations in the EC or SFG (Fig. 4c–d), or in the prefrontal cortex in Mathys et al.14 (Extended Data Fig. 7). Although Marinaro et al. reported broad depletion of inhibitory neuron subpopulations in familial monogenic AD, there was no strong evidence of selective vulnerability in particular inhibitory neuron subpopulations relative to other inhibitory neuron subpopulations in Marinaro et al.
Fig. 4 |. Inhibitory neuron subpopulations do not consistently show differences in resilience or vulnerability to AD progression.
a-b, tSNE projection of inhibitory neurons from the EC (a) and SFG (b) in their respective alignment spaces, colored by individual of origin (center) or subpopulation identity (outer). c-d, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations across Braak stages (middle subpanel); for each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of inhibitory neuron molecular subtype markers from Lake et al.12 (bottom subpanel).
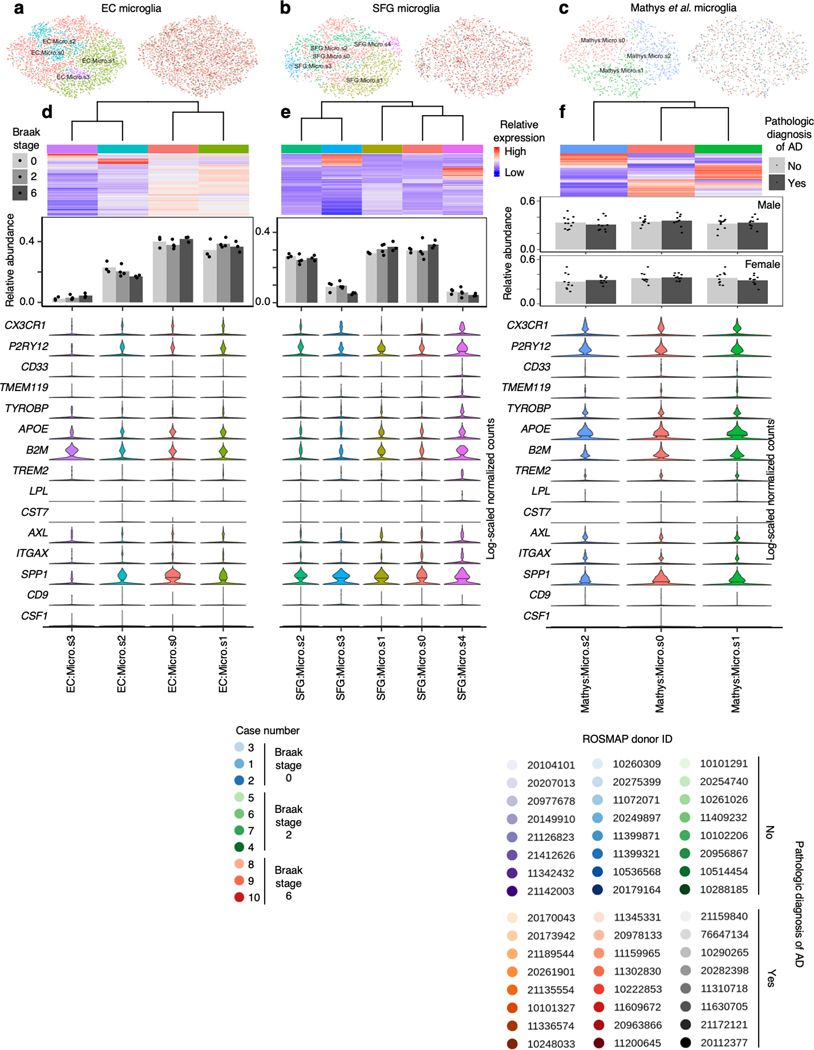
Analysis of glial subpopulations
Glial cells have emerged as important players in AD. We found a trend towards increased relative abundance of microglia in the EC in with AD progression (Fig. 1f), consistent with microgliosis. Next, we asked whether a specific transcriptional state of microglia is associated with AD in our dataset. Recent single-cell profiling of microglia from mouse models of AD identified disease-associated microglia30 (DAM), the transcriptional signature of which overlap only partially with that of human microglia found in AD31. Considering the possibility that DAMs may cluster separately from homeostatic microglia after cross-sample alignment, we performed subclustering of microglia in our dataset, discerning 4 subpopulations in the EC and 5 subpopulations in the SFG (Extended Data Fig. 8a–b). However, similar to Thrupp et al.32, we were unable to detect the expression of the majority of homeostatic microglia markers and DAM markers in our dataset or in Mathys et al.14 (Extended Data Fig. 8d–f), which may be due to the relatively low number of genes captured in microglia compared to other cell types (Fig. 1h–i) and the depletion of many DAM markers in nuclei compared to whole cells32.
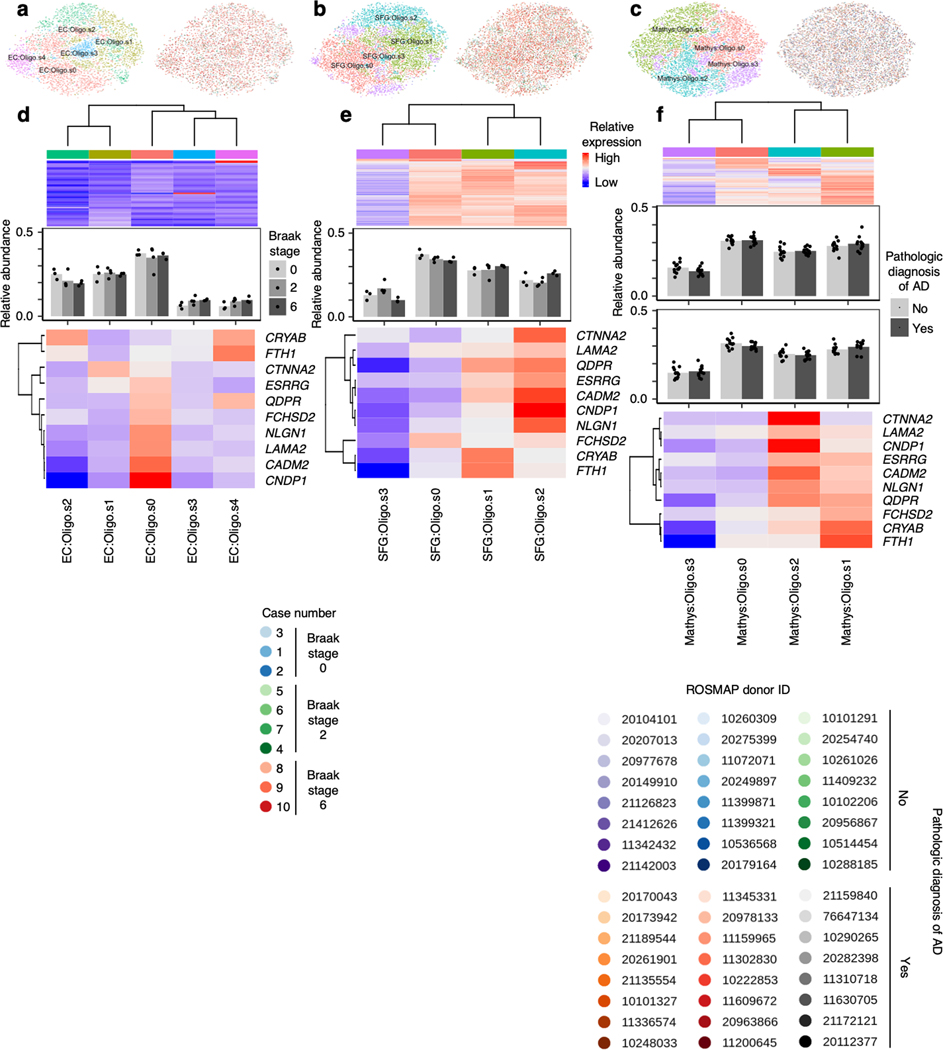
We next examined oligodendrocytes, which were shown by Mathys et al.14 to exhibit a strong transcriptional response in AD. Subclustering of oligodendrocytes in the EC and SFG revealed subpopulations (EC:Oligo.s0 and EC:Oligo.s4, SFG:Oligo.s1 and SFG:Oligo.s2) which exhibited higher expression of AD-associated oligodendrocyte genes from Mathys et al.14, i.e. genes with higher expression in the AD-associated subpopulation Oli0 in Mathys et al.14 (Extended Data Fig. 9d–e). Although the function of these genes in the context of AD is largely unknown, a spatial transcriptomics study of AD33 has recently implicated a subset of these genes in the response of oligodendrocytes to amyloid plaques (e.g. CRYAB, QDPR).
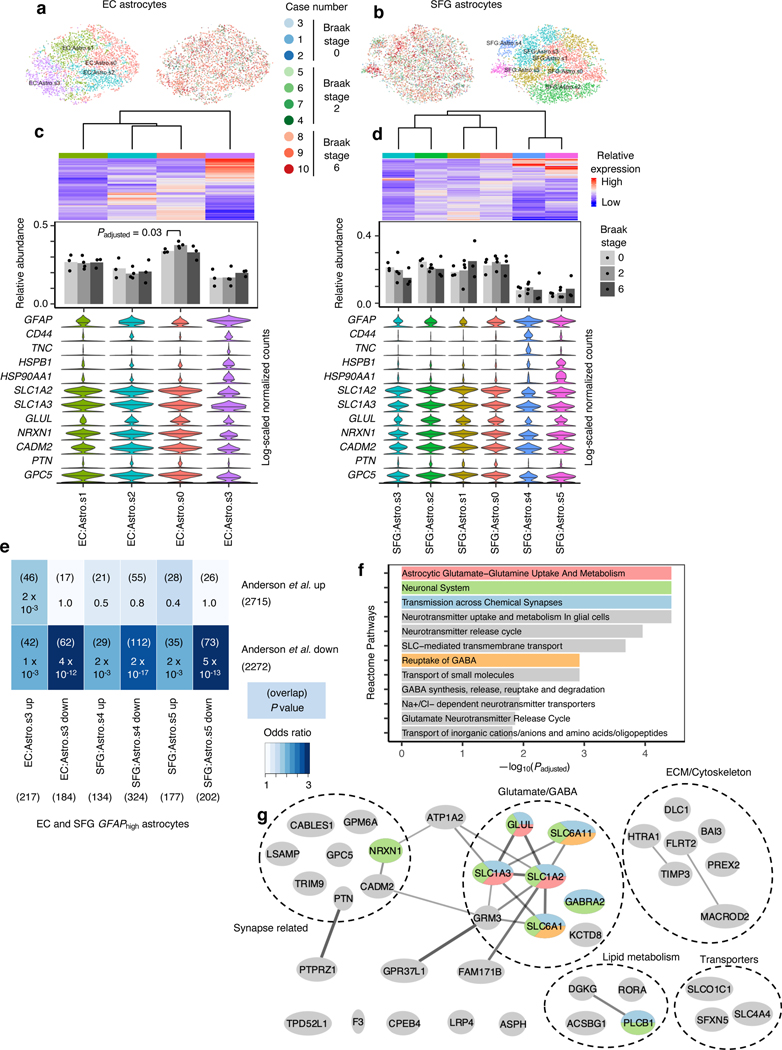
Finally, we turned our attention to astrocytes. While reactive astrocytes are ubiquitously associated with AD pathology34, only few studies to date have directly profiled reactive astrocytes due to the difficulty of specifically isolating reactive astrocytes35, 36. Similarly to our interrogation of microglia, we asked if reactive astrocytes would cluster separately from non-reactive astrocytes after cross-sample alignment. After subclustering of astrocytes in our dataset, we discerned 4 subpopulations in the EC and 6 subpopulations in the SFG (Fig. 5a–d). In each brain region, there was at least one subpopulation (EC:Astro.3, SFG:Astro.s4 and SFG:Astro.s5) that expressed dramatically higher levels of GFAP, which we will refer to as GFAPhigh astrocytes (Fig. 5c,d). In the EC, GFAPhigh astrocytes also expressed CD44 and HSPB1, markers of pan-reactive astrocytes37; TNC, which is upregulated in stab-wound reactive astrocytes38; and HSP90AA1, which is upregulated in reactive astrocytes associated with middle cerebral artery occlusion39 (Fig. 5c,d). Interestingly, in the SFG, GFAPhigh astrocytes consisted of two subpopulations, one marked by higher expression of CD44 and TNC, both of which are involved in interactions with the extracellular matrix, and the other marked by higher expression of HSPB1 and HSP90AA1, both of which are chaperones involved in proteostasis. In terms of downregulated genes, GFAPhigh astrocytes consistently expressed lower levels of genes associated with glutamate/GABA homeostasis (SLC1A2, SLC1A3, GLUL, SLC6A11) and synaptic adhesion/maintenance (NRXN1, CADM2, PTN, GPC5), suggesting a loss of homeostatic function.
Fig. 5 |. GFAPhigh astrocytes show signs of dysfunction in glutamate homeostasis and synaptic support.
a-b, tSNE projection of astrocytes from the EC (a) and SFG (b) in their respective alignment spaces, colored by individual of origin (center) or subpopulation identity (outer). c-d, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). ). Relative abundance of subpopulations across Braak stages (middle subpanel); for each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression of genes associated with reactive astrocytes, with median expression level marked by line (bottom subpanel). e, Enrichment analysis of overlap between differentially expressed genes in GFAPhigh astrocytes vs. differentially expressed genes in reactive astrocytes from Anderson et al.40 The number of genes in each gene set and the number of overlapping genes are shown in parentheses, and the hypergeometric test p-values (one-sided, corrected for multiple testing using the Benjamini-Hochberg procedure) are shown without parentheses. f, Enrichment of Reactome pathways in downregulated genes in GFAPhigh astrocytes, with selected terms highlighted in color. g, Functional association network (see Methods) of downregulated genes shared between EC and SFG GFAPhigh astrocytes that overlap with those in Anderson et al.40 Genes with stronger associations are connected by thicker lines. Genes that belong to selected gene sets in f are highlighted in color.
Examination of all differentially expressed genes in GFAPhigh astrocytes showed significant overlap with differentially expressed genes from reactive astrocytes in a mouse model of spinal cord injury40 (Fig. 5e). Overlapping downregulated genes included the previously noted genes associated with glutamate homeostasis and synaptic adhesion/maintenance and also genes related to lipid metabolism, cytoskeleton and extracellular matrix, and transporters (Fig. 5f–g).
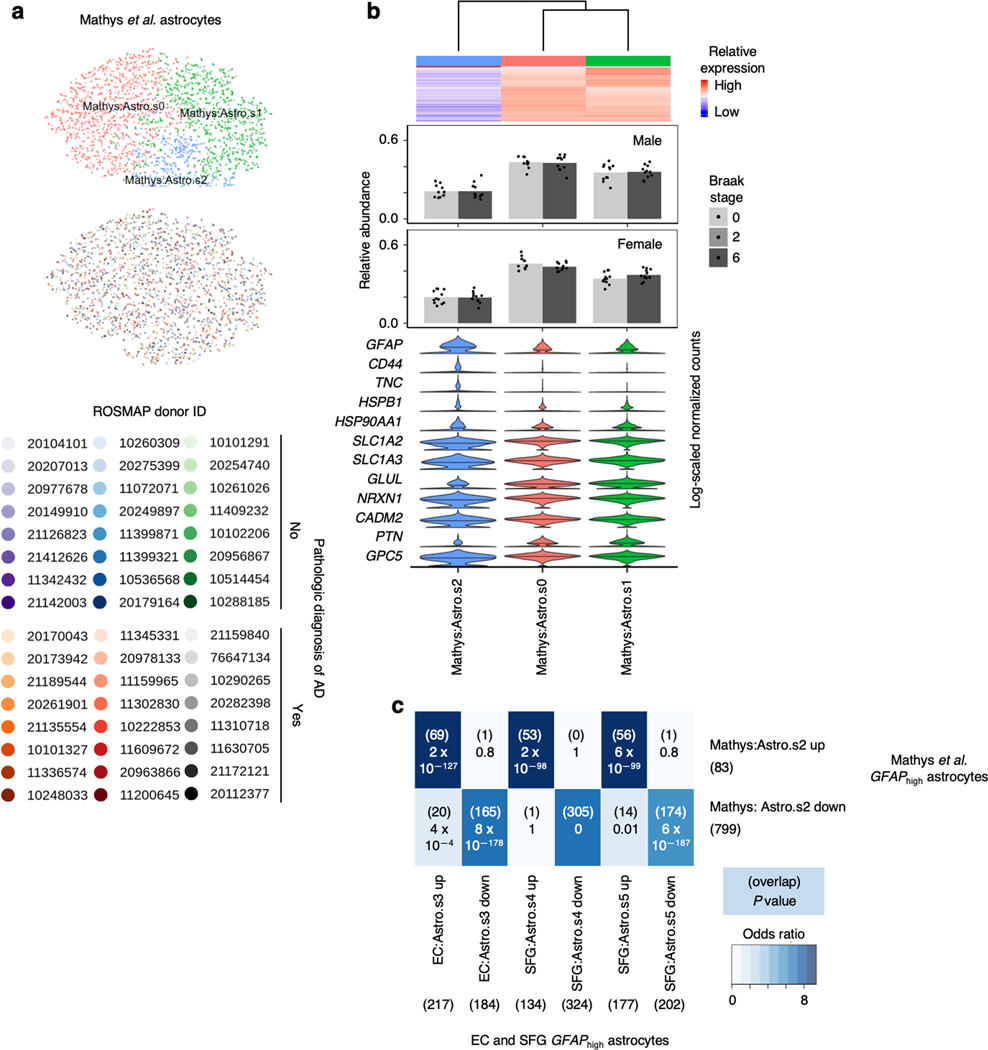
Finally, to confirm the presence of GFAPhigh astrocytes in an independent dataset, we performed subclustering of astrocytes from Mathys et al.14 after cross-sample alignment, which yielded 3 subpopulations (Extended Data Fig. 10a,b). Indeed, we found that Mathys:Astro.s2 behaved identically compared to GFAPhigh astrocytes in our dataset in terms of upregulating reactive astrocyte markers and downregulating genes associated with glutamate/GABA homeostasis and synaptic adhesion (Extended Data Fig. 10b). Furthermore, the differentially expressed genes in Mathys:Astro.s3 overlapped highly with those in GFAPhigh astrocytes in our dataset (Extended Data Fig. 10c).
DISCUSSION
Selective vulnerability is a fundamental feature of neurodegenerative diseases, including AD. Past studies have characterized the most vulnerable neurons in AD based on topography and morphology. For instance, EC layer II neurons are more vulnerable than EC layer III pyramidal neurons8–10. However, the molecular signature of selectively vulnerable neurons in AD is largely unknown.
Using a combination of snRNA-seq and quantitative neuropathology in postmortem human brains, we discovered that in the caudal EC, specific excitatory neuron subpopulations defined by snRNA-seq were selectively vulnerable in AD. These neurons expressed genes associated with layer II of the mouse medial EC, consistent with the known vulnerability of neurons in superficial layers of the human EC in AD5–8. .We identified and validated RORB as a marker of these selectively vulnerable excitatory neuron subpopulations. Selectively vulnerable RORB+ excitatory neurons included both large multipolar neurons and pyramidal neurons. Our findings demonstrates that morphology alone is insufficient to determine selective vulnerability.
We also found that tau neuronal inclusions, a chief AD neuropathological hallmark, preferentially accumulated in RORB+ excitatory neurons in the EC. To uncover potential cell biological mechanisms underlying the vulnerability of EC RORB+ excitatory neurons, we compared the gene expression profiles of EC RORB-expressing excitatory neurons against all other EC excitatory neurons, which revealed differences in the expression of genes encoding synapse- vs. axon-localized proteins, potassium channel subunits, G-protein signaling molecules, and neurotransmitter receptor signaling molecules. Future studies utilizing in vitro and animal models of AD together with techniques for manipulating gene expression such as CRISPR inhibition and activation41–43 will make it possible to address these potential mechanistic connections among RORB-expression, phospho-tau accumulation, and vulnerability.
In neocortical areas, layers III and V are the first to accumulate tau neurofibrillary inclusions in AD1, 44, 45. Our dataset, together with our re-analysis of datasets from Mathys et al.14 and Marinaro et al.26 suggests that in the neocortex, vulnerable excitatory neuron subpopulations express RORB and have a similar transcriptional profile as selectively vulnerable neurons in the EC, although not all neocortical RORB+ neurons are vulnerable. Given that RORB is known to function as a developmental driver of neuronal subtype identity in the neocortex23–25, we hypothesize that the vulnerability of RORB-expressing excitatory neuron subpopulations in different brain regions may be caused by gene expression programs driven by RORB and potentially other subtype-determining transcription factors. Further mechanistic studies involving the perturbation of RORB expression in cell-based or animal models of AD are necessary to test this hypothesis.
A previous study suggested changes in the number of neurons expressing calbindin and parvalbumin, which tend to mark inhibitory neurons, in EC layer II in AD46. Here, we found no evidence of selective vulnerability in inhibitory neurons subpopulations in EC layer II or any other layer. Inhibitory neurons in EC superficial layers show a gradient of abundance in the various EC regions20, which could confound the results. But, given that we used strict cytoarchitectonic criteria to sample the EC, it is unlikely that our results reflect comparisons of different EC areas across the cases. Evidence suggests that these inhibitory neurons undergo changes in morphology and function, rather than loss in sporadic AD46. Thus, our results do not preclude the possibility that inhibitory neuron subpopulations may be differentially affected by AD progression at the morphological and functional level, even if neuronal loss is not apparent.
Accumulating evidence is highlighting the importance of glial changes in AD. We discovered an astrocyte subpopulation expressing high levels of GFAP, which we termed GFAPhigh astrocytes, in both EC and SFG, and in prefrontal cortex from Mathys et al.14 We found that GFAPhigh astrocytes expressed higher levels of other genes associated with reactive astrocytes, and lower levels of genes involved in astrocyte homeostatic functions. Furthermore, we found a high degree of overlap between genes differentially expressed in GFAPhigh astrocytes and genes differentially expressed in reactive astrocytes from a mouse model of spinal injury40. Thus, we believe that GFAPhigh astrocytes correspond to reactive astrocytes in AD, which may have compromised homeostatic function.
Our study has several methodological strengths. First, the postmortem cohort used for snRNA-seq and histopathological validation consists of well-characterized cases, devoid of non-AD pathology. To minimize confounders in the snRNA-seq results, we selected only male cases with an APOE ε3/ε3 genotype. Second, we sequenced a very large number of nuclei from each case (~10,000 nuclei per case, compared to ~1,700 nuclei per case in Mathys et al.14) from two brain regions per individual (~4,000 nuclei from the EC and ~6,000 nuclei from the SFG). Third, we used strict cytoarchitectonic criteria to sample brain regions for snRNA-seq and histopathological validation, instead of broadly defined sampling areas used by previous studies. Fourth, our focus was on defining cell type subpopulations that showed changes in relative abundance between disease stages, which can reflect important disease processes such as neuronal loss, and to define the genes characteristic of these subpopulations. By defining cell type subpopulations independently of disease progression, we could compare gene expression between subpopulations within individuals while controlling for differences among individuals; this is more robust than comparing gene expression in a given subpopulation across groups of individuals, which can be influenced by differences in confounding factors between the groups. Lastly, by validating our findings using a novel multiplex immunofluorescence approach29, we could quantify the relative abundance of excitatory neurons and RORB+ neurons and also demonstrate that RORB+ excitatory neurons were preferentially affected by neurofibrillary inclusions.
A limitation of our study is that we only included male APOE ε3/ε3 individuals in the snRNA-seq analysis. We included females and individuals carrying the APOE ε4 allele associated with AD risk in our histopathological validation, but caution should be taken before generalizing our results to these groups. Future studies will provide a systematic analysis of the impact of sex and APOE status on selective vulnerability in AD.
In conclusion, our study contributes a pioneering characterization of selectively vulnerable neuronal populations in AD using snRNA-seq profiling of paired brain regions from the same individuals, which were all carefully curated AD cases and controls. These results will inform future studies of the mechanistic basis of selective vulnerability in both animal and in vitro models, such as human iPSC-derived neurons, in which the deployment of CRISPR inhibition and activation technology enables elucidation of the functional consequences of transcriptomic changes41, 47.
ONLINE METHODS
Post-mortem cohort
This study was approved by and University of Sao Paulo institutional review board and deemed non-human subject research by the University of California, San Francisco (UCSF). De-identified human postmortem brain tissue was supplied by the Neurodegenerative Disease Brain Bank (NDBB) at UCSF, and the Brazilian BioBank for Aging Studies (BBAS) from the University of Sao Paulo50. The NDBB receives brain donations from patients enrolled in the UCSF Memory and Aging Center research programs. The BBAS is population‐based and houses a high percentage of pathologically and clinically normal control subjects who are not available in the NDBB. Neuropathological assessments were performed using standardized protocols and followed internationally accepted criteria for neurodegenerative diseases51–53. The brain samples used in this study contained a broad burden of AD-type pathology and were selected to be free from non-AD pathology including Lewy body disease, TDP-43 proteinopathies, primary tauopathies, and cerebrovascular changes. Argyrophilic grain disease (AGD) was not an exclusion criterion based on its high prevalence and lack of correlation with significant clinical symptoms54–56. In total, the cohort included 10 cases who underwent snRNA-seq, representing Braak stages 0, 2 and 6, all ApoE 3/3, and 26 cases who underwent neuroanatomical analysis, representing Braak stages 0–61, 57, ranging from 2–5 individuals per Braak stage. Table 1 depicts the characteristics of the 31 cases.
Isolation of nuclei from frozen post-mortem human brain tissue
Isolation of nuclei was performed similarly as previously described58. Briefly, frozen brain tissue was dounce homogenized in 5 ml of lysis buffer (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tricine-KOH, pH 7.8, 1 mM DTT, 0.15mM spermine, 0.5 mM spermidine, 1X protease inhibitor (Sigma, 4693159001), and RNAse Inhibitor (Promega, N2615)). Following initial dounce homogenization, IGEPAL-630 was added to a final concentration of 0.3% and the sample was homogenized with 5 more strokes. The solution was then filtered through a 40 um cell filter and mixed with Optiprep (Sigma, D1556–250ML) to create a 25% Optiprep solution. This solution was then layered onto a 30%/40% Optiprep gradient and centrifuged at 10,000g for 18 minutes using the SW41-Ti rotor. The nuclei were collected at the 30%/40% Optiprep interface.
Droplet-based single-nucleus RNA-sequencing
Droplet-based single-nucleus RNA-sequencing (snRNA-seq) was performed using the Chromium Single Cell 3′ Reagent Kits v2 from 10X Genomics. Nuclei were resuspended to a concentration of 1000 nuclei/uL in 30% Optiprep solution before loading according to manufacturer’s protocol, with 10,000 nuclei recovered per sample as the target. cDNA fragment analysis was performed using the Agilent 4200 TapeStation System. Sequencing parameters and quality control were performed as described by The Tabula Muris Consortium59.
Pre-processing of snRNA-seq data
Sequencing data generated from snRNA-seq libraries were demultiplexed using Cellranger (version 2.1.0) cellranger mkfastq. To align reads, we first generated our own pre-mRNA GRCh38 reference genome using cellranger mkref in order to account for introns that may be eliminated using the default GRCh38 reference genome. Alignment and gene expression quantification was then performed using cellranger count with default settings.
Exploratory analysis of EC and SFG data
For each sample, the raw gene-barcode matrix outputted by Cellranger was converted into a SingleCellExperiment (SCE) object in R (version 3.5.1) using the read10xCounts function from the DropletUtils package60 (version 1.2.2). Droplets containing nuclei were then distinguished from empty droplets using DropletUtils::emptyDrops with the parameter FDR = 0.01, and then nuclei (hereon also referred to as “cells”) with less than 200 UMIs were discarded. Afterwards, SCE objects corresponding to each sample were merged into a single SCE object for downstream processing and analyses.
For normalization of raw counts, to avoid artifacts caused by data sparsity, the approach of Lun et al.61 was adopted: For each sample, cells were first clustered using a graph-based method followed by pooling counts across cells in each cluster to obtain pool-based size factors, which were then deconvoluted to yield cell-based size factors. Clustering was performed using the quickCluster function from the scran package62 (version 1.10.2) with the parameters method = ‘igraph’, min.mean = 0.1, irlba.args = list(maxit = 1000), and the block parameter set to a character vector containing the sample identity of each cell. Size factors were computed using scran::computeSumFactors with the parameter min.mean = 0.1 and the cluster parameter set to a character vector containing the cluster identity of each cell; cells with negative size factors were removed. Normalization followed by log-transformation was then performed using the normalize function from the scater package63 (version 1.10.1).
Prior to dimensionality reduction, highly variable genes were identified for each sample separately using the approach of Lun et al.62: Each gene’s variance was decomposed into a technical and biological component. Technical variance was assumed as Poisson and modeled using scran::makeTechTrend. The mean-variance trend across genes was fitted using scran::trendVar with parameters use.spikes = FALSE and loess.args = list(span = 0.05); and the trend slot of the resulting fit object was then set to the output of scran::makeTechTrend. Biological variance was extracted from the total variance using scran::decomposeVar with the above fit object as the input. Finally, highly variable genes that were preserved across samples were identified by combining the variance decompositions with scran::combineVar, using Stouffer’s z-score method for meta-analysis (method = ‘z’), which assigns more weight to samples with more cells.
For initial data exploration, genes with combined biological variance greater than 0 were used as the feature set for dimensionality reduction by principal component analysis using scran::parallelPCA, which uses Horn’s parallel analysis to decide how many principal components to retain, with parameter approx = TRUE. Clustering was then performed on the retained principal components using the FindClusters function from the Seurat package64 (version 2.3.4) with parameter resolution = 0.8, which required conversion of SCE objects to Seurat objects using Seurat::Convert. To visualize the clusters, t-stochastic neighborhood embedding (tSNE) was performed on the retained principal components using scater::runTSNE with parameters perplexity = 30 and rand_seed = 100.
Cross-sample alignment of SFG and EC data
Initial data exploration revealed that clustering was driven by individual of origin in addition to cell type identity, which makes it difficult to analyze changes in the relative abundance or gene expression of a given cell type across disease progression or brain regions. To recover clusters defined by mainly by cell type identity, data was aligned across samples from each brain region using with scAlign65 (version 1.0.0), which leverages a neural network to learn a low-dimensional alignment space in which cells from different datasets group by biological function independent of technical and experimental factors. As noted by Johansen & Quon65, scAlign converges faster with little loss of performance when the input data is represented by principal components or canonical correlation vectors. Therefore, prior to running scAlign, the top 2000 genes with the highest combined biological variance were used as the feature set for canonical correlation analysis (CCA), which was implemented using Seurat::RunMultiCCA with parameter num.cc = 15. The number of canonical coordinates to use for scAlign was determined by the elbow method using Seurat::MetageneBicorPlot. scAlign was then run on the cell loadings along the top 10 canonical correlation vectors with the parameters options = scAlignOptions(steps = 10000, log.every = 5000, architecture = ‘large’, num.dim = 64), encoder.data = ‘cca’, supervised = ‘none’, run.encoder = TRUE, run.decoder = FALSE, log.results = TRUE, and device = ‘CPU’. Clustering was then performed on the full dimensionality of the ouptut from scAlign using Seurat::FindClusters with parameter resolution = 0.8 for the SFG and resolution = 0.6 for the EC. Clusters were visualized with tSNE using Seurat::RunTSNE on the full dimensinality of the output from scAlign with parameter do.fast = TRUE. Alignment using scAlign followed by clustering was also performed for all samples from both brain regions jointly.
To assign clusters identified in the aligned subspace generated by scAlign to major brain cell types, the following marker genes were used: SLC17A7 and CAMK2A for excitatory neurons, GAD1 and GAD2 for inhibitory neurons, SLC1A2 and AQP4 for astrocytes, MBP and MOG for oligodendrocytes, PDGFRA and SOX10 for oligodendrocyte precursor cells (OPCs), CD74 and CX3CR1 for microglia/myeloid cells, and CLDN5 and FLT1 for endothelial cells. Clusters expressing markers for more than one cell type, most likely reflecting doublets, were removed from downstream analyses.
Cell type-specific subclustering (subpopulation) analysis
To identify cell type subpopulations, cells from all samples belonging to a given major cell type were extracted for sample-level re-computation of size factors and highly variable genes. CCA was then performed using the top 1000 genes with the highest combined biological variance as the feature set, followed by alignment of the first 10 to 12 canoical coordinates with scAlign, with steps = 2500. The full dimensionality of the output from scAlign was used for subclustering (using resolution = 0.4) and tSNE. Analyzing cells from each brain region separately, marker genes for subpopulations were identified using scran::findMarkers with parameters direction = ‘up’, pval.type = ‘any’, lfc = 0.58, and the block parameter set to a character vector corresponding to each cell’s sample identity. Subpopulations that expressed markers for more than one cell type were removed from downstream analyses.
Identification of differentially expressed genes in cell type subpopulations
To identify genes differentially expressed by a cell type subpopulation compared to all other subpopulations in a way that accounts for true biological replication (i.e. at the level of individuals), UMI counts of cells from the same individual belonging to the subpopulation of interest or all other subpopulations were summed to obtain “pseudo-bulk” samples, which were then analyzed using edgeR66 (version 3.24.3) following the approach recommended by Amezquita et al.67 A false-discovery rate cutoff of 0.1 was used.
Heatmap visualization of relative gene expression across cell types or cell type subpopulations
For heatmaps of relative gene expression across cell types or cell type subpopulations shown in the figures, log-scaled normalized counts of each gene were z-score transformed across all cells and then averaged across cells in each cluster to enhance visualization of differences among clusters. Thus genes with “high” relative expression have above-average expression (positive z-scores) and genes with “low” relative expression have below-average expression (negative z-scores).
Functional association network analysis and pathway enrichment analysis of differentially expressed genes
Differentially expressed genes were visualized as a functional association network using String-db68 (v11), a protein-protein association network based on known physical interactions, functional associations, coexpression, and other metrics, and Cytoscape69 (version 3.7.2), a network visualization software. When generating the networks, the String-db association confidence score cutoff set to 0.5, and the network layout was optimized for visualization using the yFiles Organic Layout. For pathway enrichment analysis, enrichments for Gene Ontology terms and Reactome Pathways were also obtained through String-db, using a false-discovery rate cutoff of 0.05.
Entorhinal cortex layer-specific genes
Due to the lack of published data on layer-specific genes for the human EC, layer-specific genes in the mouse medial entorhinal cortex (MEC) were obtained from Ramsden et al.22. (The MEC is the most phylogenetically similar to the human caudal EC20, 21 used in this study.) Specifically, genes with expression specific for layer II, III, and V/VI of the mouse MEC according to the S4 Dataset excel spreadsheet in the supplemental information of Ramsden et al.22 were mapped to human genes, and cross-referenced against genes differentially expressed across EC excitatory neuron subclusters (obtained using scran::findMarkers without setting direction = ‘up’).
Re-analysis of the Mathys et al. dataset
To re-analyze the data from Mathys et al.14 using our cross-sample alignment approach, the filtered matrix of UMI counts (“Data/Gene Expression (RNA-seq)/Processed/filtered_count_matrix.mtx”) and associated row and column metadata (“filtered_gene_row_names.txt” and “filtered_column_metadata.txt”) were downloaded from the AMP-AD Knowledge Portal (Synapse ID: syn18485175). The experimental and clinical metadata files were downloaded from “Data/Metadata/”. The filtered UMI counts matrix and the associated row and column metadata were then converted to a SingleCellExperiment object for analysis, and the relevant experimental and clinical metadata (e.g. “Pathologic diagnosis of AD” were merged with the SingleCellExperiment object. The cell type assignments from Mathys et al.14 provided in the column metadata were used for subclustering.
Functional annotation of differentially expressed genes in GFAPhigh astrocytes
We obtained the functional annotation for differentially expressed genes from the GeneCards website70 and verified the primary literature references for glutamate/GABA-related genes71–76 and synaptic adhesion/maintenance-related genes77–80.
Quantitative histopathological assessment using multiplex immunofluorescence
Delineation of the caudal EC.
We used archival paraffin blocks from the UCSF/NBDD and BBAS (Table 1). First, we collected blocks sampling the hippocampal formation anterior to the lateral genicular body from the 10 cases used for the snRNAseq and another 30 cases spanning all Braak stages1. To determine if the caudal EC region was present, 8μm thick sections of each block underwent hematoxylin and eosin staining (Extended Data Fig. 8A). We took digital images of the stained sections and aligned each one the most approximate section from a large collection of 400 μm thick serial coronal sections of whole-brain hemispheres stained for gallocyanin provided by co-author Heinsen19, 81 (Extended Fig Data 8B). We eliminated blocks from five cases used for snRNA-seq and four of the extra cases for lack of caudal EC. Next, again with the aid of the paired gallocyanin sections, we delineated the borders of the caudal EC in each case (Extended Data Fig. 8A).
The EC is considered a peri- or allocortex, depending on the author9. EC parcellation and cytoarchitectonic definitions have been a matter of debate, and here, we are adopting the cytoarchitectonic definitions proposed by Heinsen and colleagues19, which is based on the examination of thick histological preparations and considered the definitions proposed by Insausti and Amaral (6 layers)82 and Braak and Braak (3 layers)9. In thick histological sections, the caudal entorhinal region features well-delineated clusters of stellate or principal cells in layer II (pre-alpha clusters) and three lamina dissecans19. The external dissecans (dissecans-ext) divides layers II and III is particularly prominent in the caudal EC. Dissecans-1 (diss-1) corresponds to layer IV of Insausti83 and the lamina dissecans of Braak and Braak9 and Rose84. The most internal dissecans (dissecans-2, or diss-2) is hardly appreciated in thin sections but easy to visualize in thick sections. It roughly corresponds to layer Vc of the caudal subregions of Insausti83.
Multiplex immunofluorescence.
Next, for each case, an 8μm thick, formalin-fixed and paraffin-embedded coronal section underwent immunofluorescence against TBR1, RORB and phospho-tau(CP-13) as described below. TBR1, or T-box, brain, 1 is a transcription factor protein that has a role in differentiation of glutamatergic neurons and is a marker for excitatory neurons, including EC excitatory neurons85, 86. In summary, sections were deparaffinized and incubated in 3.0% hydrogen peroxide (Fisher, H325–500) in methanol to inactivate endogenous peroxidase. Antigen retrieval was performed in 1X Tris-EDTA HIER solution (TES500) PBS with 0.05% Tween 20 (PBS-T) at pH9 in an autoclave at 121 °C for five minutes. To reduce nonspecific background staining, sections were blocked with 5% Milk/PBS-T. To avoid cross-reactions between primary antibodies that were raised against the same species, an antibody stripping step using 0.80% β-mercaptoethanol/10% sodium dodecyl sulfate in 12.5% Tris-HCL was performed after the tyramide-signal amplification (TSA) development for RORB.
Sections were first incubated overnight in primary antibody against RORB (1:400, rabbit, HPA008393, Millipore Sigma), which was later developed in goat anti-rabbit HRP (1:400, R-05072–500, Advansta) with Alexa Fluor 488 TSA (1:100, B40953, Thermo Fisher). Next, sections were stripped of RORB primary antibody and then were incubated overnight in a cocktail of primary antibodies against TBR1 (1:100, Rabbit, ab31940, Abcam) and CP13 (1:800, mouse, phospho-tau serine 202, gift of Peter Davies, NY), all of which were later developed with secondary antibodies and fluorophores: for TBR1, Alexa Fluor 546 conjugated anti-rabbit secondary (1:200, A-11010, Thermo Fisher) was used, and for CP13, biotinylated anti-mouse (1:400, BA-2000, Vector Laboratory) with streptavidin Alexa Fluor 790 (1:250, S11378, Thermo Fisher) was used. Sections were then counterstained with DAPI diluted in PBS (1:5000, D1306, Invitrogen). Finally, sections were then incubated in Sudan Black B (199664–25g, Sigma) to reduce autofluorescence and coverslipped using Prolong antifade mounting media (P36980, Invitrogen). A quality control slide was used to verify the efficacy of the antibody stripping process. A detailed description of the method is provided in Ehrenberg et al.29 Sections were scanned using a Zeiss AxioScan Slide Scanner.
For generating the images shown in Fig. 3h, a section from case #6 (Braak stage 2, see Table 1) was stained with gallocyanin-chrome alum following standard methods19. The section was placed on a cover slip and scanned using a Zeiss AxioScan Slide Scanner. Next, the section was removed from the cover slip and underwent immunofluorescence for RORB and CP13 as described above. Then, the section was placed on a cover slip and scanned once more.
Neuronal quantification.
The caudal EC delineations carried out in the hematoxylin and eosin-stained slides were then transferred to the immunostained images. Within these borders, we randomly placed four 500×500 μm regions of interest (ROI) overlaying the EC external layers (I to III), which we identified as being external to dissecans-1. We then extracted the ROIs for quantification in ImageJ (Fig. 3). The number of excitatory neurons was quantified by segmenting the TBR1 signal, using a threshold to create a mask and the segmentation editor plugin to manually remove all non-neuronal artifacts and vessels. The number of RORB+ excitatory neurons was then counted using the mask of excitatory (TBR1+) neurons in the segmentation editor and manually removing all neurons not expressing RORB. All segmentations were manually verified for quality control. Quantification was done blinded to the neuropathological diagnosis. We quantified phospho-tau (CP-13) staining in two ROIs in a subset of the cases, using the same FIJI protocol.
Statistics
Beta regression.
For each brain region, the relative abundance of a given cell cluster or cell type, which ranges from 0 to 1, was computed for each sample, treated as an independent measurement, and assumbed to follow a beta distribution (although this was not formally tested). To determine the statistical significance of changes in the relative abundance of a given cluster or cell type across Braak stages, beta regression87 was performed using the betareg package (version 3.1–1), using the formula relative.abundance ~ braak.stage for both the mean and precision models, and the bias-corrected maximum likelihood estimator (type = ‘BC’). The statistical significance of changes in the proportion of TBR1+ cells and RORB+ cells among TBR1+ cells obtained from immunofluorescence validation were assessed similarly as above using beta regression. To correct for multiple hypothesis testing for each family of tests (e.g. testing all cell type subpopulations for a brain region), Holm’s method was used to adjust P values obtained from beta regression to control the family-wise type I error rate at 0.05.
Fisher’s exact test.
For Fig. 3g, the two-sided Fisher’s Exact Test was used to calculate the statistical significance of the observed enrichment of CP13 staining in RORB+ excitatory neurons. The test was performed in R using fisher.test with alternative=‘two-sided’.
Hypergeometric test.
For Fig. 5e and Extended Data Fig. 10c, the one-sided hypergeometric test (implemented in R with the package GeneOverlap, version 1.18.0) was used to calculate the statistical significance of the observed gene overlaps. The P values were adjusted for multiple testing using the Benjamini-Hochberg method.
Randomization.
Data collection for the snRNA-seq or immunostaning validation was not randomized or blocked.
Sample sizes.
No statistical methods were used to pre-determine sample sizes but our sample sizes are comparable to those reported in previous publications14, 15.
A Life Sciences Reporting Summary is provided as Supplementary information.
Extended Data
Extended Data Fig. 1. Data quality and initial clustering without cross-sample alignment.
a-b, Mean number of genes (a) or UMIs (b) detected per cell across individual samples for major cell types identified in each dataset. Grubman et al.15 did not resolve excitatory neurons from inhibitory neurons. Pericytes were identified only in Mathys et al.14 Cell type abbreviations: Exc – excitatory neurons, Oligo – oligodendrocytes, Astro – astrocytes, Inh – inhibitory neurons, OPC – oligodendrocyte precursor cells, Micro – microglia, Endo – endothelial cells, Per – pericytes. c-d, tSNE projection of cells from the EC (c) and SFG (d) clustered without first performing cross-sample alignment, colored by individual of origin (center) or cluster assignment (outer). e-f, Heatmap and hierarchical clustering of clusters and cluster marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Expression of cell type markers (bottom subpanels).
Extended Data Fig. 2. Expression of selected EC excitatory neuron subpopulation markers and pathway enrichment analysis of differentially expressed genes in selectively vulnerable EC excitatory neuron subpopulations.
a, Expression heatmap of genes that are specifically expressed by four or fewer EC excitatory neuron subpopulations; “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). b-d, Enrichment analysis against Gene Ontology Cellular Component terms or Reactome Pathways (b,d) and functional association network analysis (c,e; see Methods) of genes with higher (b-c) or lower expression (d-e) in RORB+ vulnerable EC excitatory neurons, with selected terms highlighted by color. In panels c and e, genes with stronger associations are connected by thicker lines, and genes without known associations are not shown.
Extended Data Fig. 3. Differential expression analysis across Braak stages for EC excitatory neuron subpopulations.
a-b, Number of differentially expressed genes in EC excitatory neuron subpopulations with higher (a) or lower (b) expression in Braak stage 6 vs. Braak stage 0. c-f, Enrichment analysis against Gene Ontology Cellular Component terms (c-d) or Reactome Pathways (e-f) of differentially expressed genes in EC excitatory neuron subpopulations with higher (c,e) or lower (d,f) expression in Braak stage 6 vs. Braak stage 0.
Extended Data Fig. 4. Alignment of EC and SFG maps homologous excitatory neuron subpopulations.
a, tSNE projection of excitatory neurons from the EC and SFG in the joint alignment space, colored by subpopulation identity (top), individual of origin (middle), or brain region (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations across Braak stages (second and third subpanels); for each brain region, statistical significance of differences in relative abundance across Braak stages (Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of EC layer-specific genes identified from Ramsden et al.22 (fourth subpanel). Expression heatmap of neocortical layer-specific genes from Lake et al.12 (fifth subpanel). Expression of selectively vulnerable EC excitatory neuron subpopulation markers by excitatory neurons in the EC (sixth subpanel) or SFG (bottom subpanel). Significant beta regression P values (adjusted for multiple testing) are shown in a table at the bottom of the panel. c, Sankey diagram connecting subpopulation identity of excitatory neurons in the EC alignment space and the SFG alignment space to subpopulation identity in the EC+SFG alignment space. The links connecting EC:Exc.s2 and EC:Exc.s4 to SFG:Exc.s2 and SFG:Exc.s4, respectively, are highlighted.
Extended Data Fig. 5. Cross-sample alignment of excitatory neurons from Mathys et al. recapitulates selective vulnerability in a RORB-expressing subpopulation.
a, tSNE projection of excitatory neurons from Mathys et al.14 in the alignment space, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (second and third subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n=12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of neocortical layer-specific genes from Lake et al.12 (fourth subpanel). Expression of selectively vulnerable EC excitatory neuron subpopulation markers (bottom subpanel). c, Heatmap of Pearson correlation between the gene expression profiles of excitatory neuron subpopulations from the EC vs. those from the prefrontal cortex in Mathys et al.14
Extended Data Fig. 6. Delineation of the EC for each case used in immunofluorescence validation.
a, The borders of the caudal EC delineated on sections stained with hematoxylin and eosin (H&E) for all 26 cases used in immunofluorescence validation (Table 1). b, Borders of the EC were determined with the aid of 400 um thick serial coronal sections of whole-brain hemispheres stained with gallocyanin (see Methods). Each H&E section (left) along with its corresponding immunofluorescence image (middle) was aligned to the most approximate gallocyanin section (right), in which the the dissecans layers (diss-1, diss-2, and diss-ext) characteristic of the caudal EC were easier to visualize. This was then used to guide delineation of the EC on the H&E and immunofluorescence sections. For more details on the cytoarchitectonic definitions used to define the caudal EC, please consult Heinsen et al.19.
Extended Data Fig. 7. Inhibitory neurons from Mathys et al. also do not show differences in resilience or vulnerability to AD.
a, tSNE projection of inhibitory neurons from Mathys et al.14 in the alignment space, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation markers (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (second and third subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n=12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression heatmap of inhibitory neuron subtype markers from Lake et al.12 (bottom subpanel).
Extended Data Fig. 8. Subclustering of microglia does not sufficiently resolve disease associated microglia signature.
a-c, tSNE projection of astrocytes from the EC (a), SFG (b), and Mathys et al.14 (c) in their respective alignment spaces, colored by subpopulation identity (left) or individual of origin (right). d-f, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations (middle subpanels) across Braak stages in the EC and SFG (for each brain region, Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) or between AD cases vs. controls in Mathys et al.14 (for each sex, cases n =12, controls n = 12, where n is the number of individuals sampled); statistical significance of differences in relative abundance was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression of disease associated microglia markers, with median expression level marked by line (bottom subpanels).
Extended Data Fig. 9. Subclustering of oligodendrocytes identifies subpopulations with higher expression of AD-associated oligodendrocyte markers from Mathys et al.
a-c, tSNE projection of oligodendrocytes from the EC (a), SFG (b), and Mathys et al.14 (c) in their respective alignment spaces, colored by subpopulation identity (left) or individual of origin (right). d-f, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanels); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations (middle subpanels) across Braak stages in the EC and SFG (for each brain region, Braak 0 n=3, Braak 2 n=4, Braak 6 n=3, where n is the number of individuals sampled) or between AD cases vs. controls in Mathys et al.14 (for each sex, cases n =12, controls n = 12, where n is the number of individuals sampled); statistical significance of differences in relative abundance was determined by beta regression and adjusted for multiple comparisons (see Methods). Relative expression of AD-associated oligodendrocyte subpopulation markers from Mathys et al.14 (bottom subpanels).
Extended Data Fig. 10. Astrocyte subpopulations with high GFAP expression from Mathys et al. are highly similar to those from the EC and SFG.
a, tSNE projection of astrocytes from Mathys et al.14 in the alignment subspace, colored by subpopulation identity (top) or individual of origin (bottom). b, Heatmap and hierarchical clustering of subpopulations and subpopulation marker expression (top subpanel); “High” and “Low” relative expression reflect above- and below-average expression, respectively (see Methods). Relative abundance of subpopulations in in AD cases vs. controls, separated by sex (middle subpanels); for each sex, statistical significance of differences in relative abundance between AD cases vs. controls (cases n=12, controls n=12, where n is the number of individuals sampled) was determined by beta regression and adjusted for multiple comparisons (see Methods). Expression of genes associated with reactive astrocytes, with median expression level marked by line (bottom subpanel). c, Enrichment analysis of overlap between differentially expressed genes in astrocytes with high GFAP expression from Mathys et al.14 vs. differentially expressed genes in astrocytes with high GFAP expression from the EC and SFG; the number of genes in each gene set and the number of overlapping genes are shown in parentheses, and the hypergeometric test p-values are shown without parentheses.
ACKNOWLEDGEMENTS
We thank A. Pisco, A. Maynard, S. Darmanis and the MACA team at the Chan Zuckerberg Biohub for advice on analysis 10X library preparation and reagents. We thank members of the Kampmann lab (A. Samelson, X. Guo, R. Tian, B. Rooney) for feedback on the manuscript. This work was supported by NIH awards F30 AG066418 (K.L.), K08 AG052648 (S.S.), R56 AG057528 (M.K., L.T.G.), K24 AG053435 (L.T.G), U54 NS100717 (L.T.G, M.K.), an NDSEG fellowship (E.L.), Alzheimer’s Association fellowship AARF 18-566005 (R.D.R.), FAPESP/CAPES (2016/24326-0) (R.D.R.) and a Chan Zuckerberg Biohub Investigator Award (M.K.). The UCSF Neurodegenerative Disease Brain Bank is supported by NIH grants AG023501 and AG019724, the Tau Consortium, and the Bluefield Project to Cure FTD.
Footnotes
DATA AVAILABILITY
The raw snRNA-seq sequencing data and unfiltered UMI count matrices are available on the Gene Expression Omnibus (GEO) under the accession GSE147528 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE147528). Single-cell data after quality control is available for download in synapse.org at under the Synapse ID syn21788402 (https://www.synapse.org/#!Synapse:syn21788402). Post- quality control data can also be explored interactively through the CellXGene platform at https://kampmannlab.ucsf.edu/ad-brain. Data from Mathys et al.14 was downloaded from Synapse under Synapse ID syn18485175 (https://www.synapse.org/#!Synapse:syn18485175).
CODE AVAILABILITY
We provide the full bioinformatics pipeline for the analysis of snRNA-seq data in this paper at https://kampmannlab.ucsf.edu/ad-brain-analysis.
ACCESSION CODES
Gene Expression Omnibus (GEO): GSE147528. Synapse: syn21788402, syn18485175.
COMPETING INTERESTS
The authors declare no competing interests.
ETHICS DECLARATIONS
This project was approved the ethical committee of the University of Sao Paulo (for tissue transfer) and deemed non-human subject research by UCSF.
Supplementary Material
REFERENCES
- 1.Braak H & Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol 82, 239–259 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Scholl M, et al. PET Imaging of Tau Deposition in the Aging Human Brain. Neuron 89, 971–982 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seeley WW, Crawford RK, Zhou J, Miller BL & Greicius MD Neurodegenerative diseases target large-scale human brain networks. Neuron 62, 42–52 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braak H & Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging 16, 271–278; discussion 278–284 (1995). [DOI] [PubMed] [Google Scholar]
- 5.Price JL, et al. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer disease. Arch Neurol 58, 1395–1402 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Stranahan AM & Mattson MP Selective vulnerability of neurons in layer II of the entorhinal cortex during aging and Alzheimer’s disease. Neural Plast 2010, 108190 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Hoesen GW, Hyman BT & Damasio AR Entorhinal cortex pathology in Alzheimer’s disease. Hippocampus 1, 1–8 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Isla T, et al. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci 16, 4491–4500 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braak H & Braak E. The human entorhinal cortex: normal morphology and lamina-specific pathology in various diseases. Neurosci Res 15, 6–31 (1992). [DOI] [PubMed] [Google Scholar]
- 10.Kordower JH, et al. Loss and atrophy of layer II entorhinal cortex neurons in elderly people with mild cognitive impairment. Ann Neurol 49, 202–213 (2001). [PubMed] [Google Scholar]
- 11.Drummond E & Wisniewski T. Alzheimer’s disease: experimental models and reality. Acta Neuropathol 133, 155–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodge RD, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathys H, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubman A, et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nature neuroscience 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Nelson PT, et al. Correlation of Alzheimer disease neuropathologic changes with cognitive status: a review of the literature. J Neuropathol Exp Neurol 71, 362–381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hof PR, et al. Parvalbumin-immunoreactive neurons in the neocortex are resistant to degeneration in Alzheimer’s disease. J Neuropathol Exp Neurol 50, 451–462 (1991). [DOI] [PubMed] [Google Scholar]
- 18.Fu H, et al. A tau homeostasis signature is linked with the cellular and regional vulnerability of excitatory neurons to tau pathology. Nature neuroscience 22, 47–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinsen H, et al. Quantitative investigations on the human entorhinal area: left-right asymmetry and age-related changes. Anat Embryol (Berl) 190, 181–194 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Kobro-Flatmoen A & Witter MP Neuronal chemo-architecture of the entorhinal cortex: A comparative review. Eur J Neurosci 50, 3627–3662 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Naumann RK, et al. Conserved size and periodicity of pyramidal patches in layer 2 of medial/caudal entorhinal cortex. J Comp Neurol 524, 783–806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsden HL, Surmeli G, McDonagh SG & Nolan MF Laminar and dorsoventral molecular organization of the medial entorhinal cortex revealed by large-scale anatomical analysis of gene expression. PLoS Comput Biol 11, e1004032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jabaudon D, Shnider SJ, Tischfield DJ, Galazo MJ & Macklis JD RORbeta induces barrel-like neuronal clusters in the developing neocortex. Cereb Cortex 22, 996–1006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oishi K, Aramaki M & Nakajima K. Mutually repressive interaction between Brn1/2 and Rorb contributes to the establishment of neocortical layer 2/3 and layer 4. Proceedings of the National Academy of Sciences of the United States of America 113, 3371–3376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa Y & O’Leary DD Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci 25, 234–244 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Marinaro F, et al. Molecular and cellular pathology of monogenic Alzheimer’s disease at single cell resolution. bioRxiv, 2020.2007.2014.202317 (2020). [Google Scholar]
- 27.Liang WS, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: a reference data set. Physiol Genomics 33, 240–256 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Franjic D, et al. Molecular Diversity Among Adult Human Hippocampal and Entorhinal Cells. bioRxiv doi: 10.1101/2019.12.31.889139 (2019). [DOI] [Google Scholar]
- 29.Ehrenberg AJ, et al. A manual multiplex immunofluorescence method for investigating neurodegenerative diseases. Journal of Neuroscience Methods In press, 10.1016/j.jneumeth.2020.108708. (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keren-Shaul H, et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Srinivasan K, et al. Alzheimer’s patient brain myeloid cells exhibit enhanced aging and unique transcriptional activation. bioRxiv doi: 10.1101/610345 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thrupp N, et al. Single nucleus sequencing fails to detect microglial activation in human tissue. bioRxiv, 2020.2004.2013.035386 (2020). [Google Scholar]
- 33.Chen WT, et al. Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer’s Disease. Cell (2020). [DOI] [PubMed] [Google Scholar]
- 34.Perez-Nievas BG & Serrano-Pozo A. Deciphering the Astrocyte Reaction in Alzheimer’s Disease. Front Aging Neurosci 10, 114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson JE, et al. Microarray analysis of the astrocyte transcriptome in the aging brain: relationship to Alzheimer’s pathology and APOE genotype. Neurobiol Aging 32, 1795–1807 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Sekar S, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging 36, 583–591 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liddelow SA, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laywell ED, et al. Enhanced expression of the developmentally regulated extracellular matrix molecule tenascin following adult brain injury. Proceedings of the National Academy of Sciences of the United States of America 89, 2634–2638 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zamanian JL, et al. Genomic analysis of reactive astrogliosis. J Neurosci 32, 6391–6410 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson MA, et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kampmann M. A CRISPR Approach to Neurodegenerative Diseases. Trends in molecular medicine 23, 483–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kampmann M. CRISPR-based functional genomics for neurological disease. Nat Rev Neurol (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian R, et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255 e212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hof PR & Morrison JH Neocortical neuronal subpopulations labeled by a monoclonal antibody to calbindin exhibit differential vulnerability in Alzheimer’s disease. Exp Neurol 111, 293–301 (1991). [DOI] [PubMed] [Google Scholar]
- 45.Hof PR, Cox K & Morrison JH Quantitative analysis of a vulnerable subset of pyramidal neurons in Alzheimer’s disease: I. Superior frontal and inferior temporal cortex. J Comp Neurol 301, 44–54 (1990). [DOI] [PubMed] [Google Scholar]
- 46.Mikkonen M, Alafuzoff I, Tapiola T, Soininen H & Miettinen R. Subfield- and layer-specific changes in parvalbumin, calretinin and calbindin-D28K immunoreactivity in the entorhinal cortex in Alzheimer’s disease. Neuroscience 92, 515–532 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Tian R, et al. CRISPR Interference-Based Platform for Multimodal Genetic Screens in Human iPSC-Derived Neurons. Neuron 104, 239–255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montine TJ, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol 123, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes CP, Berg L, Danziger WL, Coben LA & Martin RL A new clinical scale for the staging of dementia. Br J Psychiatry 140, 566–572 (1982). [DOI] [PubMed] [Google Scholar]
- 50.Grinberg LT, et al. Brain bank of the Brazilian aging brain study group - a milestone reached and more than 1,600 collected brains. Cell Tissue Bank 8, 151–162 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Hyman BT, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement 8, 1–13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suemoto CK, et al. Neuropathological diagnoses and clinical correlates in older adults in Brazil: A cross-sectional study. PLoS Med 14, e1002267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cairns NJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol 114, 5–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferrer I, Santpere G & van Leeuwen FW Argyrophilic grain disease. Brain 131, 1416–1432 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez RD & Grinberg LT Argyrophilic grain disease: An underestimated tauopathy. Dement Neuropsychol 9, 2–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez RD, et al. Argyrophilic Grain Disease: Demographics, Clinical, and Neuropathological Features From a Large Autopsy Study. J Neuropathol Exp Neurol 75, 628–635 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braak H, Thal DR, Ghebremedhin E & Del Tredici K. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol 70, 960–969 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Mo A, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tabula Muris C, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lun ATL, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome biology 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lun AT, Bach K & Marioni JC Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome biology 17, 75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lun AT, McCarthy DJ & Marioni JC A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy DJ, Campbell KR, Lun AT & Wills QF Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuart T, et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johansen N & Quon G. scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome biology 20, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amezquita RA, et al. Orchestrating single-cell analysis with Bioconductor. Nat Methods 17, 137–145 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Szklarczyk D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic acids research 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stelzer G, et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr Protoc Bioinformatics 54, 1 30 31–31 30 33 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Arriza JL, et al. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J Neurosci 14, 5559–5569 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Borden LA, et al. Cloning of the human homologue of the GABA transporter GAT-3 and identification of a novel inhibitor with selectivity for this site. Receptors Channels 2, 207–213 (1994). [PubMed] [Google Scholar]
- 73.Gendreau S, et al. A trimeric quaternary structure is conserved in bacterial and human glutamate transporters. J Biol Chem 279, 39505–39512 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Häberle J, et al. Congenital glutamine deficiency with glutamine synthetase mutations. N Engl J Med 353, 1926–1933 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Kawakami H, Tanaka K, Nakayama T, Inoue K & Nakamura S. Cloning and expression of a human glutamate transporter. Biochem Biophys Res Commun 199, 171–176 (1994). [DOI] [PubMed] [Google Scholar]
- 76.Melzer N, Biela A & Fahlke C. Glutamate modifies ion conduction and voltage-dependent gating of excitatory amino acid transporter-associated anion channels. J Biol Chem 278, 50112–50119 (2003). [DOI] [PubMed] [Google Scholar]
- 77.Südhof TC Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pellissier F, Gerber A, Bauer C, Ballivet M & Ossipow V. The adhesion molecule Necl-3/SynCAM-2 localizes to myelinated axons, binds to oligodendrocytes and promotes cell adhesion. BMC Neurosci 8, 90 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.González-Castillo C, Ortuño-Sahagún D, Guzmán-Brambila C, Pallàs M & Rojas-Mayorquín AE Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front Cell Neurosci 8, 443 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siddiqui TJ, et al. An LRRTM4-HSPG complex mediates excitatory synapse development on dentate gyrus granule cells. Neuron 79, 680–695 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Heinsen H, Arzberger T & Schmitz C. Celloidin mounting (embedding without infiltration) - a new, simple and reliable method for producing serial sections of high thickness through complete human brains and its application to stereological and immunohistochemical investigations. J Chem Neuroanat 20, 49–59 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Insausti R & Amaral DG Entorhinal cortex of the monkey: IV. Topographical and laminar organization of cortical afferents. J Comp Neurol 509, 608–641 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Insausti R, Munoz-Lopez M, Insausti AM & Artacho-Perula E. The Human Periallocortex: Layer Pattern in Presubiculum, Parasubiculum and Entorhinal Cortex. A Review. Front Neuroanat 11, 84 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rose S. Vergleichende Messungen im Allocortex bei Tier und Mensch. J Psychol Neurol 34, 250–255 (1927). [Google Scholar]
- 85.Fu H, et al. Tau Pathology Induces Excitatory Neuron Loss, Grid Cell Dysfunction, and Spatial Memory Deficits Reminiscent of Early Alzheimer’s Disease. Neuron 93, 533–541 e535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29, 353–366 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Ferrari SLP & Cribari-Neto F. Beta regression for modelling rates and proportions. J Appl Stat 31, 799–815 (2004). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.