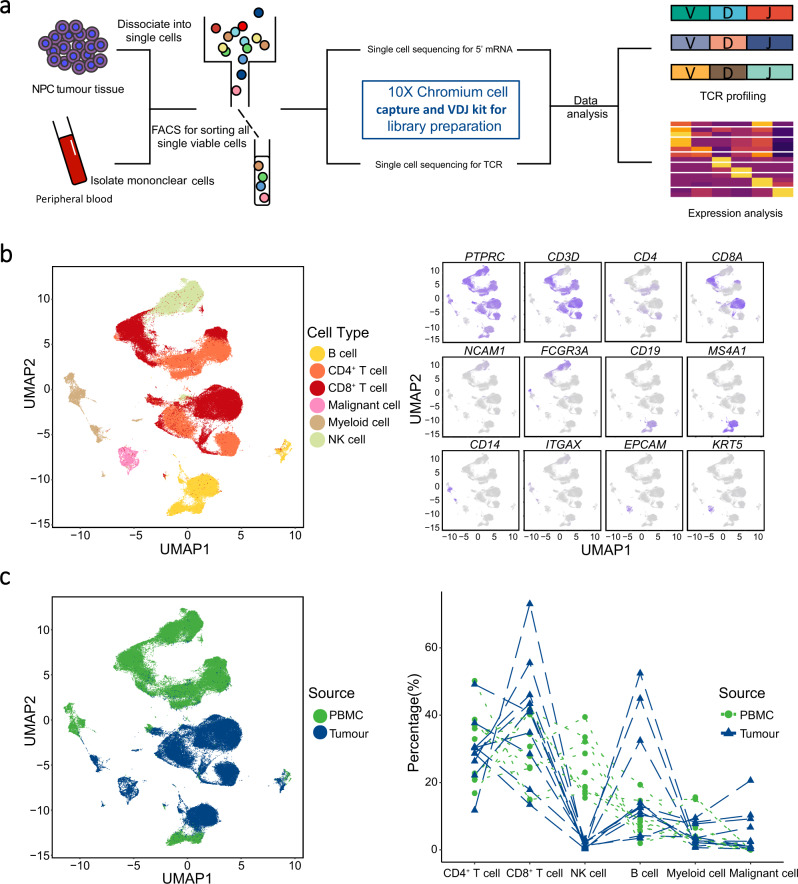
Fig. 1. The landscape profiling of single cells in NPC tumours and matching PBMC.
a An experimental scheme diagram highlights the overall study design. Single viable cells were collected using flow cytometry sorting (FACS) and subjected for cell barcoding. The cDNA libraries of 5’-mRNA expression and TCR were constructed independently, followed by high throughput sequencing and downstream analyses. b UMAP plot of 176,447 single cells grouped into six major cell types (left panel) and the normalised expression of marker genes for each cell type (right panel). Each dot represents one single cell, coloured according to cell type (left panel), and the depth of colour from grey to blue represents low to high expression (right panel). c UMAP plot of the above single cells coloured according to their origins from peripheral blood or tumour (left panel), and the fraction of cell types originating from each patient (right panel). Each dot represents one single cell, coloured according to sample origin.