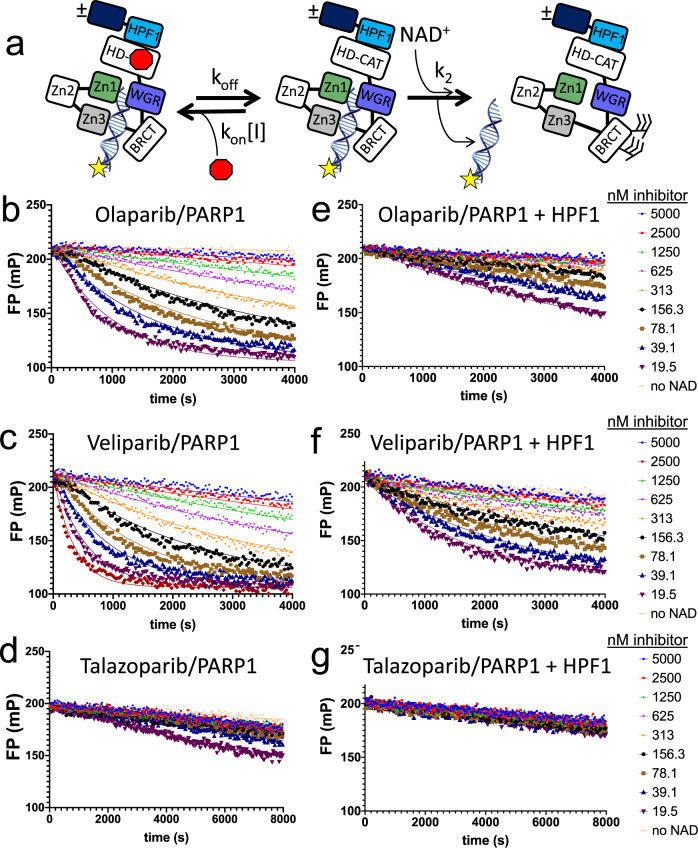
Fig. 1. Release of fluorescently labeled DNA from PARP1 reveals binding affinity of PARPi for PARP1 and demonstrates that HPF1 slows the apparent release of PARPi from PARP1.
a The domains of PARP1 involved in DNA binding (Zn1, Zn2, Zn3, WGR) and the catalytic domain with the HD motif that partially obscures the active site in the absence of DNA are shown as part of the assay scheme. The rates of association and dissociation of the inhibitor are kon and koff, respectively. The rate of PARylation of PARP1 is considered constant (k2[NAD+]) and was determined independently (see Supplementary Fig. 4). b–d Representative p18mer*-release data for olaparib, veliparib and talazoparib with PARP1 alone. e–g Representative p18mer*-release data for olaparib, veliparib and talazoparib with PARP1 in the presence of HPF1. The concentrations of the inhibitors used in (b–g) are noted on the right in units of nanomolar. The lines through the shown data points reflect fitting to first-order kinetics (see Methods).