To the Editor:
The t(10;11)(p13;q14) chromosomal translocation generating the CALM-AF10 (PICALM-MLLT10) fusion gene is found in 5–10% of T-cell acute lymphoblastic leukemias (T-ALL) and more rarely in acute myeloid leukemias (AML) [1]. These leukemias are associated with deregulation of the HOXA gene cluster and the HOXA cofactor MEIS1 [2]. Similar to MLL-AF10, CALM-AF10 binds the regulatory region of HOXA genes and activates their transcription by recruiting the H3K79 histone methyltransferase DOT1L via an octapeptide-motif/leucine zipper (OM/LZ) domain within AF10 (also called Myeloid/Lymphoid or Mixed Lineage Leukemia, translocated to chromosome 10, MLLT10) [3].
The Clathrin Assembly Lymphoid Myeloid protein CALM (or Phosphatidylinositol Binding CALM, PICALM), predominantly found in the cytoplasm, plays a role in clathrin-mediated endocytosis. We have shown that CALM contains a Nuclear Export Signal (NES) that is both necessary and sufficient for leukemic transformation of murine hematopoietic progenitor cells by enabling the binding of CALM-AF10 to Hoxa genes [4, 5]. The NES is recognized by the nuclear export protein Chromosome Regional Maintenance protein (CRM1; or Exportin 1, XPO1) which mediates the export of cargo through the nuclear pore complex (NPC), and we demonstrated by chromatin immunoprecipitation (ChIP) that CRM1 localizes at Hoxa genes [4].
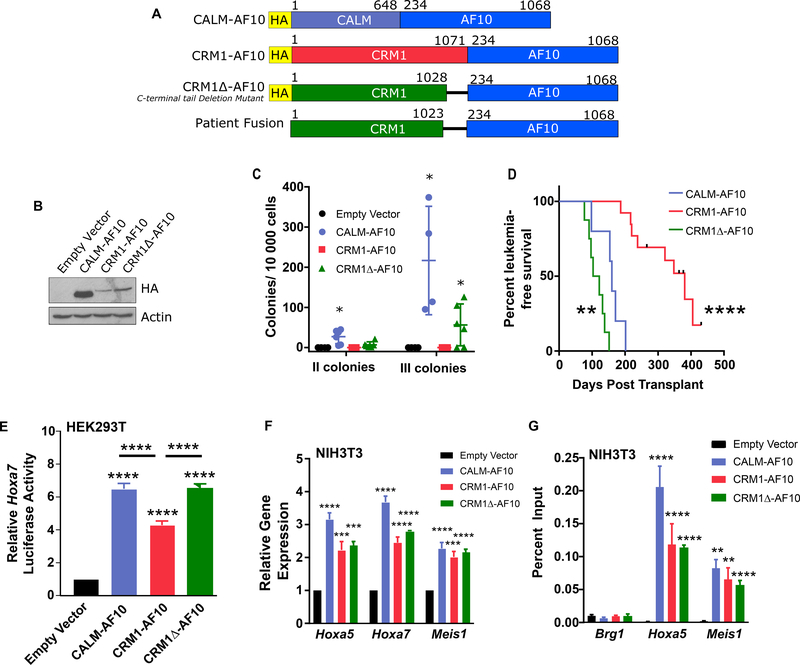
These observations support the hypothesis that an essential contribution of the CALM moiety within CALM-AF10 is to bring CRM1 in proximity to AF10. To determine whether CRM1 can substitute for CALM within the CALM-AF10 fusion, we examined the transforming properties of constructs encoding a chimeric CRM1-AF10 protein and a variant CRM1-AF10 fusion protein, CRM1Δ-AF10, that lacks the inhibitory 43 carboxy-terminal amino acid of CRM1; this construct mimics a fusion protein found in a leukemia patient (Figure 1A) [6]. Truncation of the C-terminal tail alters the conformation of CRM1 and enhances its affinity for NES-containing cargoes [7]. Both fusions displayed similar expression in transfected cells (Figure 1B). To study the transforming activity of CRM1-AF10 and CRM1Δ-AF10, we transduced hematopoietic progenitor cells (HPCs) and monitored their colony forming potential in a serial replating assay using CALM-AF10 as a positive control [5]. While cells transduced with CRM1-AF10 behaved similarly to cells transduced with the empty vector, cells transduced with CRM1Δ-AF10 exhibited enhanced clonogenic potential (Figure 1C).
Figure 1: CRM1-AF10 and CRM1Δ-AF10 display Leukemogenic Activity.
A. Schematic representation of CALM-AF10, CRM1-AF10, CRM1Δ-AF10 and the translocation fusion found in a leukemia patient [6]. The CRM1Δ-AF10 construct that we designed is truncated at amino acid 1028, whereas the CRM1-AF10 fusion isolated in the patient is truncated at amino acid 1023. Numbers above bars indicate amino acids. B. Western Blot analysis of fusion proteins. Lysates from HEK293T cells transiently transfected with the specified proteins were probed with an anti-HA antibody. CALM-AF10 is approximately 150 kDa, CRM1-AF10 and CRM1Δ-AF10 are approximately 220 kDa. C. Clonogenic assay showing the number of secondary and tertiary colonies generated from fetal liver hematopoietic cells transduced with Empty Vector, CALM-AF10, CRM1-AF10 or CRM1Δ-AF10 following serial replating in methylcellulose. Horizontal bars indicate mean number of colonies, with error bars indicating SD. One-way ANOVA test was used to determine significance compared to Empty Vector, *p<0.05. D. Kaplan-Meier curve of mice transplanted with bone marrow progenitors transduced with CALM-AF10 (n=5), CRM1-AF10 (n=13), or CRM1Δ-AF10 (n=8). Tick marks on CRM1-AF10 survival curve indicate censored mice that died without signs of leukemia. Statistical significance was determined by log-rank test, survival of CRM1-AF10 and CRM1Δ-AF10 mice was compared to CALM-AF10 mice. E. Transcriptional activation of the Hoxa7 reporter in HEK293T cells transiently co-transfected with Empty Vector, CALM-AF10, CRM1-AF10, or CRM1Δ-AF10. Luciferase values are shown relative to empty vector. Asterisks over each fusion protein indicate significance compared to empty vector. Asterisks over horizontal bars indicate significance between noted fusion proteins. Error bars indicate SD, a minimum of 15 independent experiments (transduction replicates) were measured per sample. F. Expression levels of Hoxa5, Hoxa7, and Meis1 transcripts in NIH3T3 cells stably transduced with Empty Vector, CALM-AF10, CRM1-AF10, or CRM1Δ-AF10. Values were measured by qRT-PCR and normalized to Gapdh. Results are normalized to empty vector control. Error bars indicate S.E.M., from at least 3 biological replicates. Significance is shown in comparison with empty vector. G. Binding of CALM-AF10, CRM1-AF10, and CRM1Δ-AF10 to Hoxa5 and Meis1 genes determined by ChIP analysis. The Brg1 gene is used as a negative control. ChIP was performed using an anti-HA antibody to precipitate the HA-tagged fusion proteins expressed in stably transduced NIH3T3 cells. Error bars indicate S.E.M. from at least three biological replicates, significance is shown in comparison with empty vector. E-G: Differences determined using unpaired t-tests. **p<0.01; ***p<0.001, ****p<0.0001
To study leukemogenesis, mice were transplanted with HPCs transduced with fusions using a bicistronic MSCV-IRES-eGFP vector in order to track GFP-positive transduced cells. Both cohorts of CRM1-AF10 (n=13) and CRM1Δ-AF10 (n=8) transplanted mice initially displayed similar percentages of GFP-expressing peripheral blood leukocytes, indicating comparable engraftment (Supplementary Figure 1A). Mice were observed for 450 days, during which 69% of CRM1-AF10 mice developed myeloid leukemia with a median survival of 348 days post-transplant, while 100% of CRM1Δ-AF10 mice developed leukemia with a median survival of 112.5 days post-transplant. In comparison, mice transplanted with CALM-AF10 transduced progenitors developed leukemia within 98 to 200 days (median survival 160 days) (Figure 1D). Of note, mice transplanted with HPCs transduced with either full length wild type CRM1 (n=5) or truncated CRM1Δ (aa 1–1028) (n=5) never developed leukemia (>570 days of observation, data not shown). Both CRM1-AF10 and CRM1Δ-AF10 leukemia mice showed signs of hyperleukocytosis, including splenomegaly (Supplementary Figure 1B) and invasion of bone marrow with GFP-positive leukemia blasts expressing the myeloid markers Mac-1 and Gr-1 (Supplementary Figure 1C). As we have reported for CALM-AF10 leukemia [5], CRM1-AF10 and CRM1Δ-AF10 blasts rarely expressed the pan-B lymphoid marker B220. Leukemic bone marrow blasts from CALM-AF10, CRM1-AF10 or CRM1Δ-AF10 mice expressed similar levels of Hoxa and Meis1 transcripts (Supplementary Figure 1D). Secondary leukemias could be induced by transplanting leukemic blasts recovered from either CRM1-AF10 or CRM1Δ-AF10 leukemic mice (Supplementary Figure 2).
We next assessed the ability of the fusion proteins to activate the transcription of Hoxa and Meis1 genes. Similarly to CALM-AF10, CRM1-AF10 and CRM1Δ-AF10 both stimulated expression of a Hoxa7-Luciferase reporter in transiently transfected HEK293T cells, and increased expression of endogenous Hoxa and Meis1 genes in stably transduced murine fibroblasts (Figure 1E–F). Chromatin immunoprecipitation (ChIP) assays showed that the fusion proteins bind the chromatin of these genes, indicating direct activation of their expression (Figure 1G).
We have demonstrated that a CRM1-AF10 fusion protein phenocopies CALM-AF10 in its ability to bind and activate Hoxa and Meis1 genes and to induce leukemia, supporting a model in which CRM1 enables the tethering of CALM-AF10 to Hoxa and Meis1 effector genes, thereby recruiting the AF10/DOT1L transcriptional complex and causing leukemia. We further showed that CRM1Δ-AF10, a fusion remarkably similar to that found in a T-cell leukemia patient, was superior to CRM1-AF10 in its ability to phenocopy CALM-AF10. The molecular basis for the difference in activity between CRM1Δ-AF10 and CRM1-AF10 is uncertain: as a result of the fusion, the bulky AF10 moiety (836 amino acids) is joined to the C-terminal end of CRM1, and as such, it is not clear whether removal of the inhibitory CRM1 C-terminal helix from the CRM1-AF10 fusion alters the conformation of the NES binding cleft.
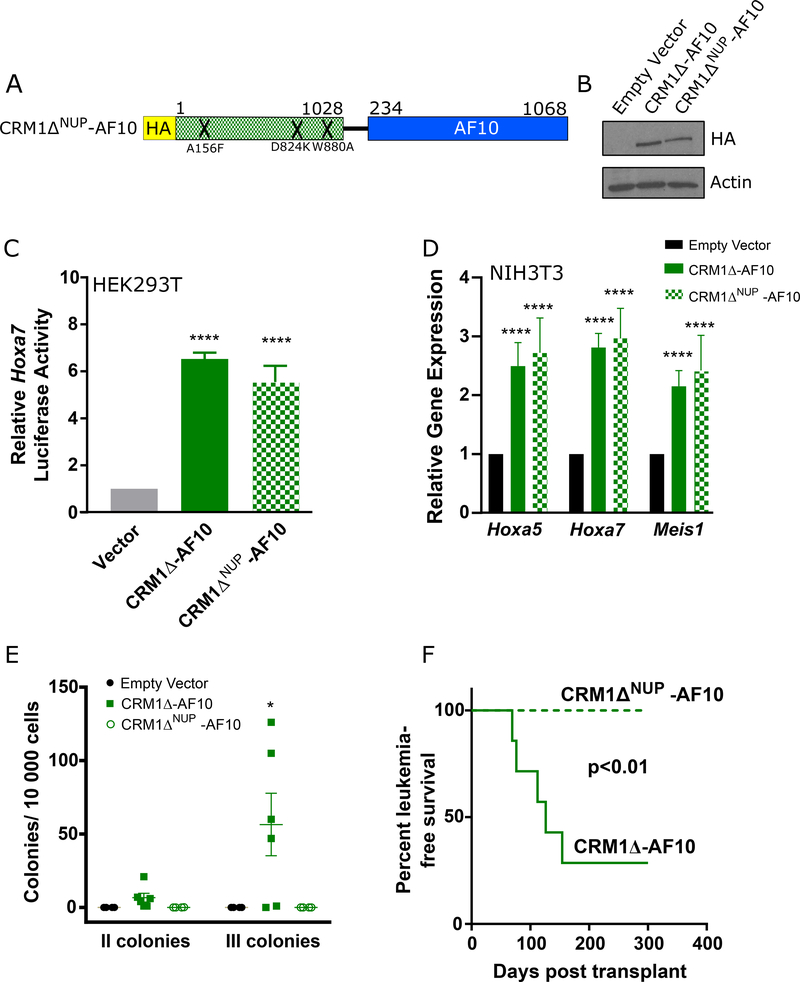
During nuclear export through the nuclear pore, CRM1 interacts with nucleoporins that contain phenylalanine-glycine repeats (FG-NUPs). Because of the involvement of FG-NUPs, such as NUP98 and NUP214, in translocations associated with leukemias that overexpress HOXA genes, we sought to explore whether the NUP-binding ability of CRM1-AF10 plays a role in leukemogenesis. The crystal structure of a CRM1/NUP214 complex identified key CRM1 residues that interact with FG motifs [8]. Mutation of three of these residues (A156F/D824K/W880A) decreases the affinity of CRM1 for NUP214 without altering the nuclear export function of CRM1 [8]. We introduced these mutations into CRM1Δ-AF10 and studied the resulting CRM1ΔNUP-AF10 mutant to investigate whether interaction with NUP214 contributes to the leukemogenic properties of CRM1Δ-AF10 (Figure 2A). We first noted that protein expression and transcriptional activity in a Hoxa7-Luciferase reporter assay of CRM1ΔNUP-AF10 was comparable to that of CRM1Δ-AF10 (Figure 2B, 2C). The effect of CRM1ΔNUP-AF10 on the expression of endogenous Hoxa genes in fibroblasts was also similar to that of CRM1Δ-AF10 (Figure 2D). In contrast, following transduction of fetal HPCs, we found that, unlike CRM1Δ-AF10, CRM1ΔNUP-AF10 did not enhance the self-renewal of colony forming cells (Figure 2E). Upon transplantation into syngeneic mice, progenitor cells transduced with CRM1Δ-AF10 or CRM1ΔNUP-AF10 initially yielded similar proportions of transduced leukocytes in the peripheral blood (i.e. expressing GFP, Supplementary Figure 3), indicating comparable engraftment. However, compared with the cohort of CRM1Δ-AF10 mice in which 5 of 7 mice developed leukemias within 45 to 154 days, none of the CRM1ΔNUP-AF10 mice (n=7) showed signs of leukemia during a 300-day observation period (Figure 2F). Using a proximity-based biotin labelling assay, we confirmed the loss of interaction of CRM1ΔNUP-AF10 with NUP214 compared with CRM1Δ-AF10; this also revealed decreased interaction of CRM1ΔNUP-AF10 with another FG-NUP, NUP98 (Supplementary Table 1). These findings suggest that while the residues involved in the interaction of CRM1Δ-AF10 with FG-NUPs are not required for the transcriptional activation of Hoxa genes, they are critical for leukemogenicity of the fusion protein.
Figure 2. Impairing the NUP214/CRM1 Interaction Abrogates Leukemia Development.
A. Schematic representation of the mutant CRM1ΔNUP-AF10 fusion protein. Mutations are indicated by an ‘X’ at the approximate location within the CRM1 moiety of the fusion protein. Numbers above bars indicate amino acids. B. Western Blot analysis of fusion proteins. Lysates from HEK293T cells transiently transfected with the specified proteins were analyzed with an anti-HA antibody. C. Transcriptional activation of the Hoxa7 reporter in HEK293T cells transiently co-transfected with Empty Vector, CRM1Δ -AF10, or CRM1ΔNUP-AF10. Luciferase values are shown relative to empty vector. Results of CRM1ΔNUP-AF10 and CRM1Δ-AF10 luciferase assays are not significantly different. Error bars indicate SD, a minimum of 5 replicates per sample were analyzed. Unpaired t-test was used to measure significance. ****p<0.0001. D. Hoxa5, Hoxa7, and Meis1 transcript levels determined by qRT-PCR in NIH3T3 fibroblasts stably expressing Empty Vector, CRM1Δ-AF10 or CRM1ΔNUP-AF10. Results were normalized to empty vector control. Error bars indicate S.E.M., from at least 3 biological replicates. Unpaired t-test was used to compare CRM1Δ-AF10 and CRM1ΔNUP-AF10 cells to empty vector controls. There were no significant differences in gene expression between CRM1Δ-AF10 and CRM1ΔNUP-AF10, ****p<0.0001. E. Clonogenic assay showing the number of secondary and tertiary colonies generated by fetal liver hematopoietic cells transduced with Empty Vector, CRM1Δ-AF10, or CRM1ΔNUP-AF10 and serially replated in methylcellulose. Lines indicate average number of colonies ± S.E.M. One-way ANOVA was used to determine significance compared to Empty Vector. *p<0.05. F. Kaplan-Meier survival curve of mice transplanted with fetal liver progenitors transduced with CRM1Δ-AF10 (n=7) or CRM1ΔNUP-AF10 (n=7). Log rank test was used to calculate p-value.
Several explanations could account for the discrepancy between the ability of CRM1ΔΔNUP-AF10 to activate transcription of Hoxa genes and its lack of leukemogenicity: leukemogenesis may be mediated by yet to be determined critical genes other than Hoxa genes; the molecular determinants for the activation of Hoxa genes may be different in leukemia precursor cells than in fibroblasts or in the Hoxa7 reporter assay; alternatively, the interaction of CRM1ΔNUP-AF10 with NUP214, or other FG-NUPs, may only be partially reduced and the threshold that triggers a reduction of Hoxa activation may be different in leukemia-initiating cells than in HEK293T cells or murine fibroblasts.
In summary, our studies suggest a critical and previously unappreciated role for CRM1 in leukemogenesis. While the CRM1-AF10 fusion is a rare occurrence in leukemia patients, we hypothesize that CRM1 may mediate leukemogenic properties of other oncoproteins causing upregulation of HOXA cluster genes, among other genes. These include oncogenic fusion proteins that interact with CRM1 via a NES, such as CALM-AF10, NAP1L1-AF10, DDX3X-AF10 or NPM1c [5, 9, 10]. CRM1 has also been recently found to be an essential cofactor for SETBP1-induced leukemias [11]. Our finding that leukemia development is abrogated upon mutation of CRM1 residues that mediate interaction with NUP214 within CRM1-AF10 suggests a role for nuclear pore proteins in CRM1-mediated leukemogenesis. While further mechanistic studies are needed to understand the role of FG-NUPs in the pathogenesis of CRM1-mediated leukemias, our findings corroborate previous observations by us and others regarding a requirement for CRM1 in HOXA activation and/or leukemogenic properties of the FG-NUP fusion oncoproteins NUP98-HOXA9 [12], SET-NUP214 [13], and SQSTM1-NUP214 [14]. The critical role of the CRM1/FG-NUP interaction could have therapeutic implications as Small Inhibitors of Nuclear Export (SINEs), which are being evaluated in multiple clinical trials including ones in leukemia patients [15], can indirectly reduce the interaction of CRM1 with FG-NUPs [8, 12].
Supplementary Material
Acknowledgements
CRM1 plasmids containing mutants in the NUP214 binding region were generously provided by Ralph Kehlenbach and Sarah Port. Pritha Bagchi, PhD and the Emory Integrated Proteomics Core provided assistance with BioID2 Mass Spectrometry. This work was supported by an American Society of Hematology Research Training Award for Fellows (WKA), Hyundai Hope on Wheels Young Investigator Award (WKA and CPL), Hyundai Hope On Wheels Scholar Award (CPL and DSW), the Duke Cancer Institute (CPL), a NIH R03 grant (1R03CA191983–01A1, CPL), Alex’s Lemonade Stand Young Investigator Award (JLH), Pablove Foundation (JLH), Pediatric Cancer Research Foundation (JLH), NHLBI T32 5T32HL007057–37 (WKA), NHLBI T32 5T32HL007057–40 (SKS), a St. Baldrick’s Foundation Research Award (DSW), and the Schiffman Family Foundation. CPL is an INSERM scientist.
Footnotes
Competing Interests Statement
None of the authors has any direct or indirect commercial financial incentive associated with publishing this article. None of the authors has an affiliation with any organization that, to our knowledge, has a direct interest in the subject matter discussed. The Wechsler Laboratory received financial support from Karyopharm, Inc. several years prior to performing the work described in this manuscript, but no Karyopharm products were used for the studies described here.
References
- 1.Kumon K, Kobayashi H, Maseki N, Sakashita A, Sakurai M, Tanizawa A, et al. Mixed-lineage leukemia with t(10;11)(p13;q21): an analysis of AF10-CALM and CALM-AF10 fusion mRNAs and clinical features. Genes Chromosomes Cancer. 1999;25(1):33–9. [DOI] [PubMed] [Google Scholar]
- 2.Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, et al. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19(11):1948–57. [DOI] [PubMed] [Google Scholar]
- 3.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–78. [DOI] [PubMed] [Google Scholar]
- 4.Conway AE, Haldeman JM, Wechsler DS, Lavau CP. A critical role for CRM1 in regulating HOXA gene transcription in CALM-AF10 leukemias. Leukemia. 2015;29(2):423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conway AE, Scotland PB, Lavau CP, Wechsler DS. A CALM-derived nuclear export signal is essential for CALM-AF10-mediated leukemogenesis. Blood. 2013;121(23):4758–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond J, Bergon A, Durand A, Tigaud I, Thomas X, Asnafi V, et al. Cryptic XPO1-MLLT10 translocation is associated with HOXA locus deregulation in T-ALL. Blood. 2014;124(19):3023–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong X, Biswas A, Chook YM. Structural basis for assembly and disassembly of the CRM1 nuclear export complex. Nat Struct Mol Biol. 2009;16(5):558–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Port SA, Monecke T, Dickmanns A, Spillner C, Hofele R, Urlaub H, et al. Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export. Cell Rep. 2015;13(4):690–702. [DOI] [PubMed] [Google Scholar]
- 9.Meijerink JP, Cante-Barrett K, Vroegindeweij E, Pieters R. HOXA-activated early T-cell progenitor acute lymphoblastic leukemia: predictor of poor outcome? Haematologica. 2016;101(6):654–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falini B, Martelli MP, Bolli N, Bonasso R, Ghia E, Pallotta MT, et al. Immunohistochemistry predicts nucleophosmin (NPM) mutations in acute myeloid leukemia. Blood. 2006;108(6):1999–2005. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen N, Oakley K, Han Y, Kwok M, Crouch G, Du Y. Interaction with XPO1 is essential for SETBP1 to induce myeloid transformation. Leukemia. 2019;33(11):2758–62. [DOI] [PubMed] [Google Scholar]
- 12.Oka M, Mura S, Yamada K, Sangel P, Hirata S, Maehara K, et al. Chromatin-prebound Crm1 recruits Nup98-HoxA9 fusion to induce aberrant expression of Hox cluster genes. Elife. 2016;5:e09540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka M, Mura S, Otani M, Miyamoto Y, Nogami J, Maehara K, et al. Chromatin-bound CRM1 recruits SET-Nup214 and NPM1c onto HOX clusters causing aberrant HOX expression in leukemia cells. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavau CP, Aumann WK, Sze SK, Gupta V, Ripple K, Port SA, et al. The SQSTM1-NUP214 fusion protein interacts with Crm1, activates Hoxa and Meis1 genes, and drives leukemogenesis in mice. PLoS One. 2020;15(4):e0232036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang AY, Liu H. The past, present, and future of CRM1/XPO1 inhibitors. Stem Cell Investig. 2019;6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.