FIGURE 2. Model for prokaryotic MMR.
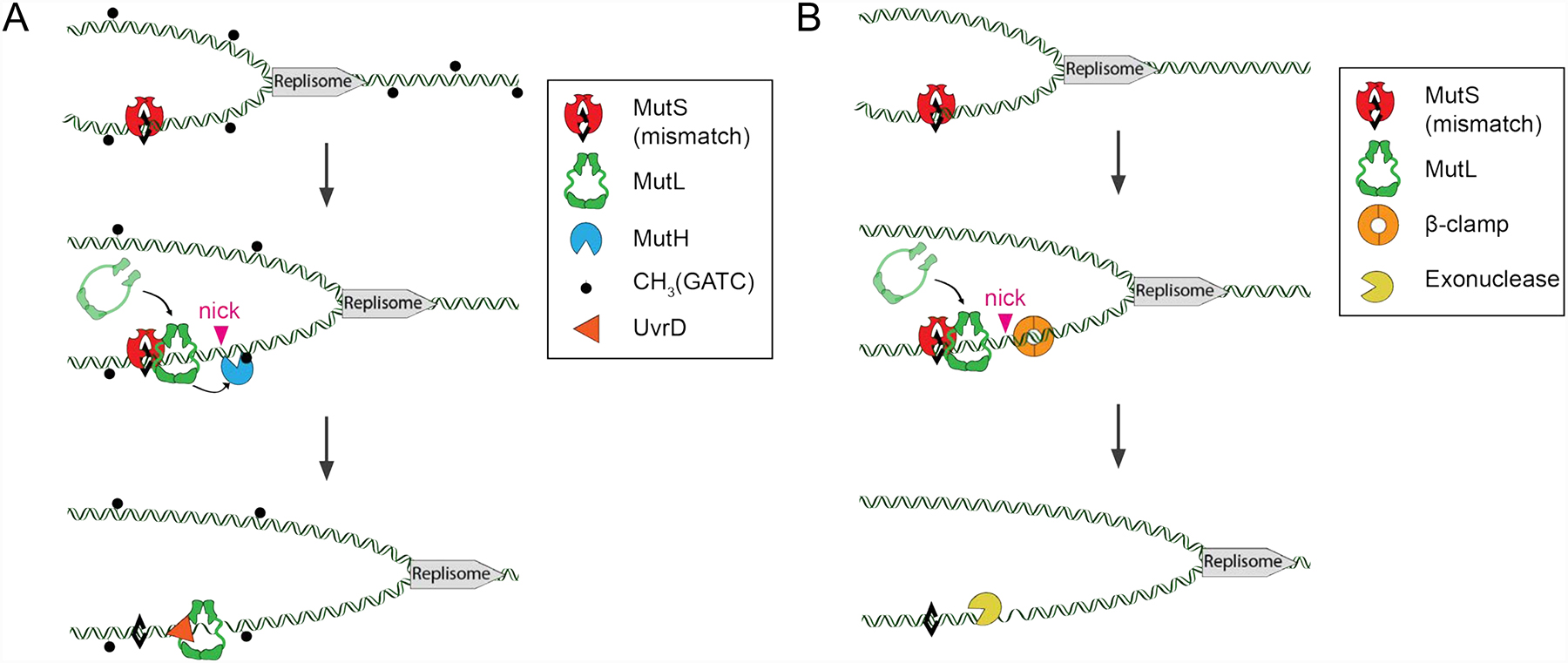
A model for mismatch repair in bacteria. A mismatch in DNA formed due to DNA polymerase misincorporation/DNA slippage is shown as a black diamond. MutS binds to the mismatch and undergoes an ATP-dependent conformational change to act as a sliding clamp and recruit MutL. (A) In E. coli, the MutS-MutL complex then acts as a sliding clamp complex that recruits MutH to nick the unmethylated (newly replicated) strand at hemi-methylated d(GATC) sites. These nicks allow for the helicase UvrD to unwind the DNA which is then excised, by a variety of nucleases, depending on the polarity of the nick relative to the mismatch. Alternatively, UvrD and MutL processively unwind the DNA between two d(GATC) sites (depicted). The resulting gap is then filled in by DNA polymerase. (B) In most other bacteria, MutL contains an intrinsic endonuclease activity that is stimulated by the DNA replication processivity factor β-clamp. These nicks act as entry sites for exonucleases to excise the mismatch, followed by re-synthesis of the gapped DNA by DNA polymerase. It is not clear if a UvrD helicase acts during MMR in bacteria that do not use MutH and Dam methylase. In such bacteria a recent model suggests that analogous to eukaryotic MMR, an exonuclease acts on double-stranded DNA to excise the mismatch, thus not requiring a UvrD helicase type activity (as shown, see Lenhart, Pillon, Guarne, Biteen, & Simmons, 2016).
