FIGURE 3. Model for MMR in baker’s yeast.
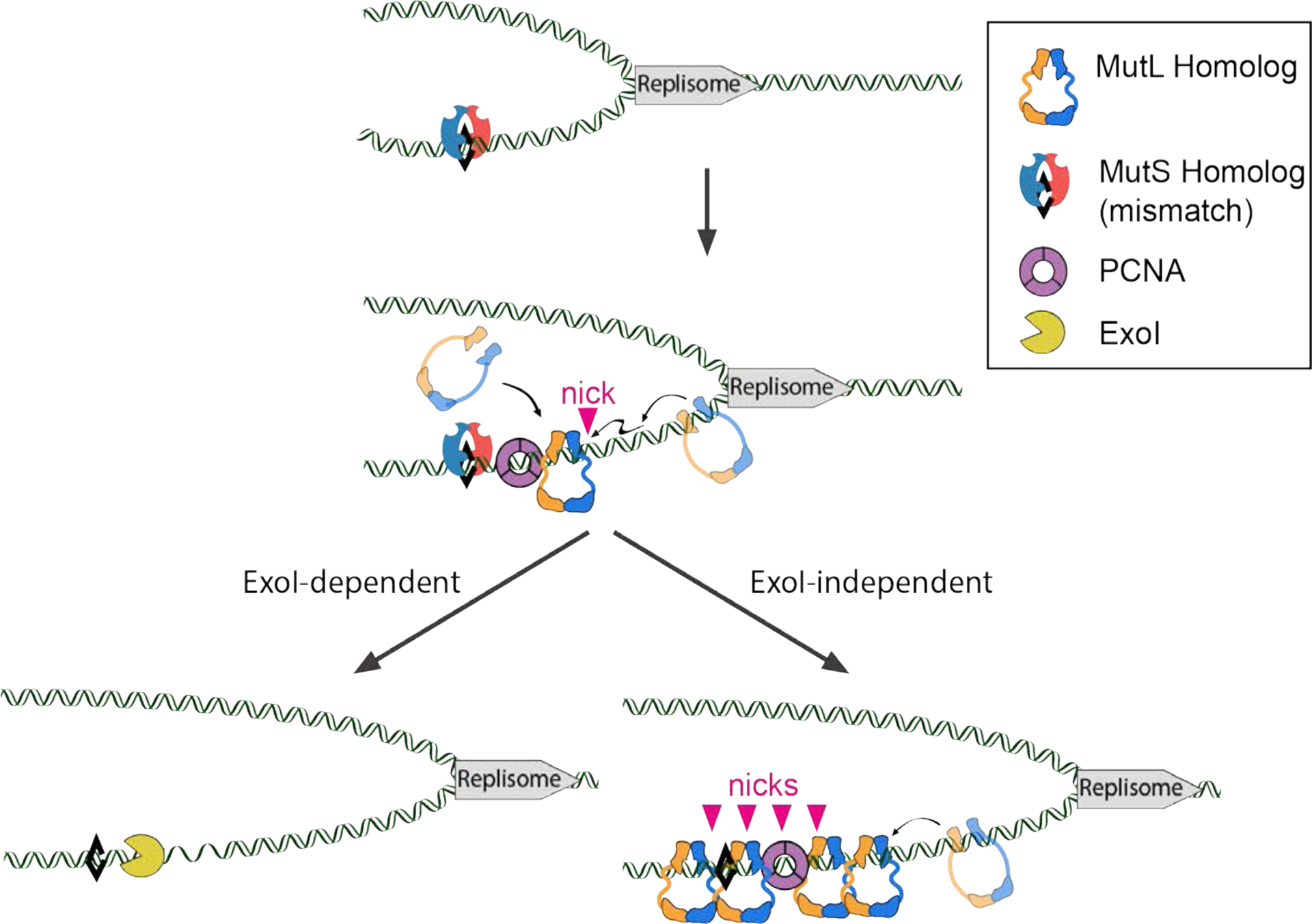
A mismatch in DNA formed due to DNA polymerase misincorporation is shown as black diamond. MSH complexes (Msh2-Msh3 or Msh2-Msh6) bind to the mismatch and undergo an ATP-dependent conformational change that allows them to act as a sliding clamp and recruit MLH complexes. Mlh1-Pms1 or Mlh1-Mlh3 undergo ATP-dependent conformational changes and orientation-specific interactions with PCNA (see text for a review of hypotheses for how these interactions with PCNA could be initiated), resulting in incision of the newly replicated DNA strand. This is followed by two potential pathways, ExoI-independent and -dependent, that can recruit downstream repair factors to excise and resynthesize the DNA and ligate the repaired strand. In the ExoI-dependent pathway, ExoI uses the nicks made by MLH complexes as an entry site to excise the mismatch containing DNA in a 5’−3’ direction. In the ExoI-independent pathway, multiple nicks made by MLH complexes in the vicinity of the mismatch promote DNA strand displacement and synthesis by DNA polymerase to repair the mismatch. Adapted from Kim et al., (2019).
