FIGURE 4. Structural homology of yeast Mlh1-Pms1, Mlh1-Mlh2, and Mlh1-Mlh3.
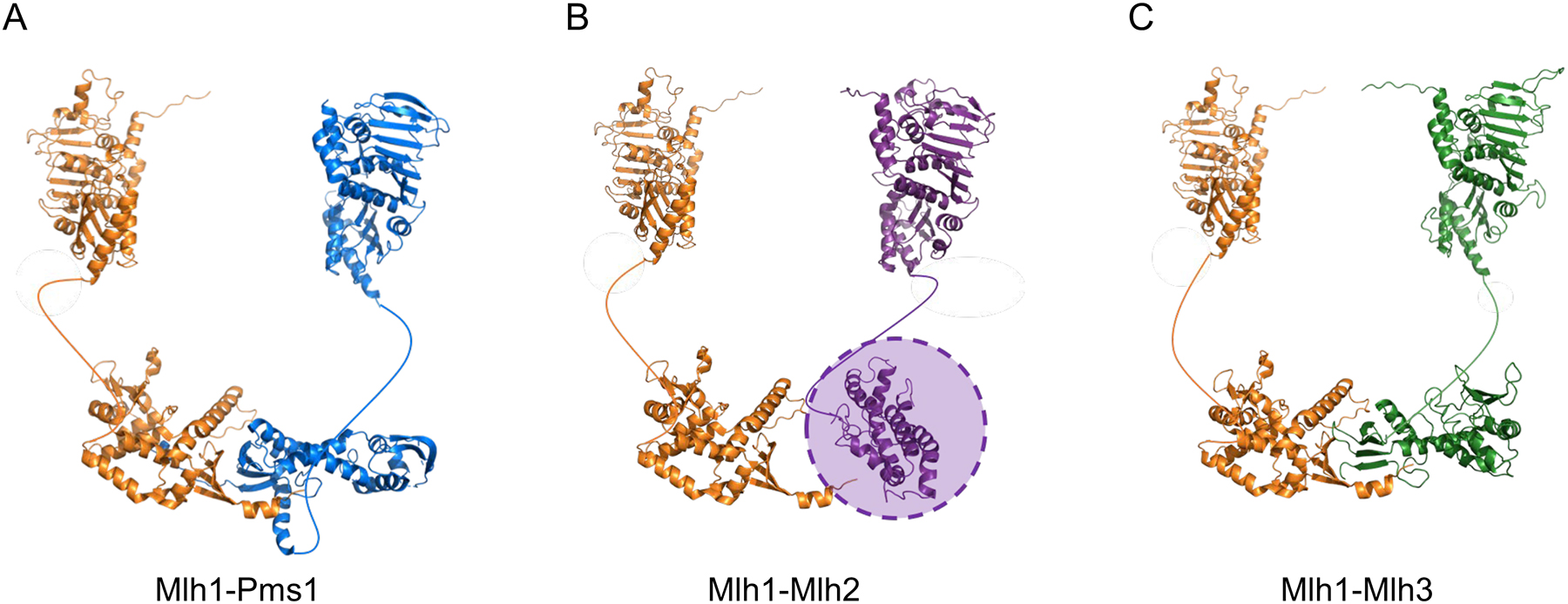
(A) For yeast Mlh1-Pms1, the N-terminal domain of Pms1 was obtained from PDB 3h4l, and the N-terminal domain of Mlh1 was modeled by a Phyre2 (amino acids 1–367 of Mlh1; http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index). The structure of the Mlh1-Pms1 C-terminus (PDB 4e4w) was solved by Gueneau et al. (2013). (B) For yeast Mlh1-Mlh2, the N-terminal domains of Mlh1 and Mlh2 were created using a Phyre2 homology model (amino acids 1–371 of Mlh2). The C-terminal domain of Mlh1 was obtained from PDB 4e4w. The C-terminal domain of Mlh2 (amino acids 503–695), created using a Phyre2 homology model, did not align well with the C-terminal domains of the other MLH proteins; for this reason, it is indicated by a shaded oval. (C) For yeast Mlh1-Mlh3, the N-terminal domains of Mlh1 and Mlh3 were created by a homology model presented in Al-Sweel et al. (2017). The C-terminal domain of Mlh1 was obtained from PDB 4e4w, and the C-terminal domain of Mlh3 (amino acids 491–715 of Mlh3) was created by a homology model presented in Al-Sweel et al. (2017). In all panels the unstructured linker domains are shown as curved lines.
