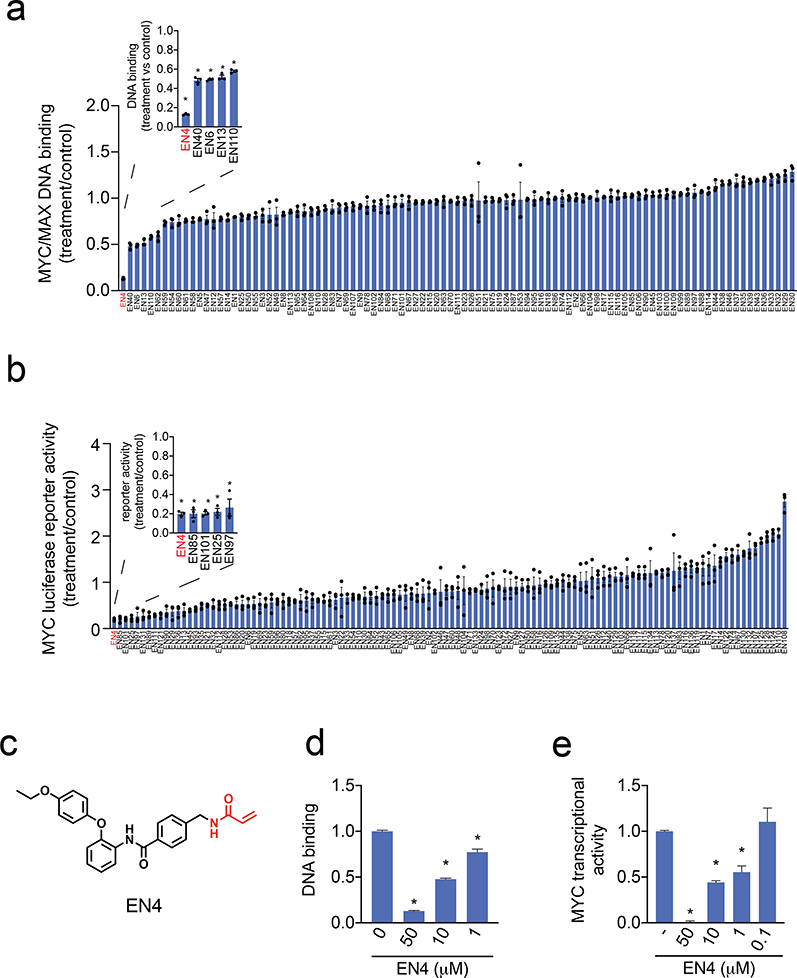
Figure 1. Covalent ligand screen to identify compounds that inhibit MYC activity in vitro and in situ.
(a) Screening a cysteine-reactive covalent ligand library in vitro for compounds that would inhibit MYC/MAX binding to its E-box DNA consensus sequence. DMSO vehicle or covalent ligands (50 μM) were pre-incubated with pure human full-length MYC protein for 30 min before direct addition of MAX and then addition to DNA binding plates for 1 h. Data are shown as ratio relative to DMSO vehicle treated controls set to 1. Shown in red was the top hit. (b) MYC luciferase reporter activity in HEK293T cells transfected with a MYC luciferase reporter construct encoding a Firefly luciferase reporter gene under the control of a basal promoter element, the TATA box, joined to tandem repeats of the MYC transcriptional E-box response element, alongside a constitutively expressing Renilla luciferase for normalization purposes. DMSO vehicle or cysteine-reactive covalent ligands (50 μM) were treated in HEK293T cells transfected with a MYC luciferase reporter gene for 24 h and Firefly and Renilla lucierase activity was read-out. Firefly luciferase levels were normalized to Renilla luciferase levels and then normalized to vehicle-treated controls. Data are shown as ratios relative to DMSO vehicle treated controls. Shown in red was the top hit. (c) Structure of top hit in both in vitro and in situ assays in (a) and (b) EN4. Shown in red is the cysteine-reactive acrylamide. (d) Dose-response of EN4 inhibition of MYC/MAX binding to its DNA consensus sequence. (e) Dose-response of EN4 inhibition of MYC luciferase reporter activity in cells. DMSO vehicle or EN4 was treated in HEK293T cells transfected with a MYC luciferase reporter gene for 24 h and luciferase activity was read-out. Data are shown as ratio relative to DMSO vehicle treated controls set to 1. Data shown in (a, b, d, e) are average ± sem, n=3 biologically independent samples/group. Statistical significance was calculated with unpaired two-tailed Student’s t-tests in (d, e) and is expressed as *p<0.05. This figure is related to Figure S1 and Table S1.