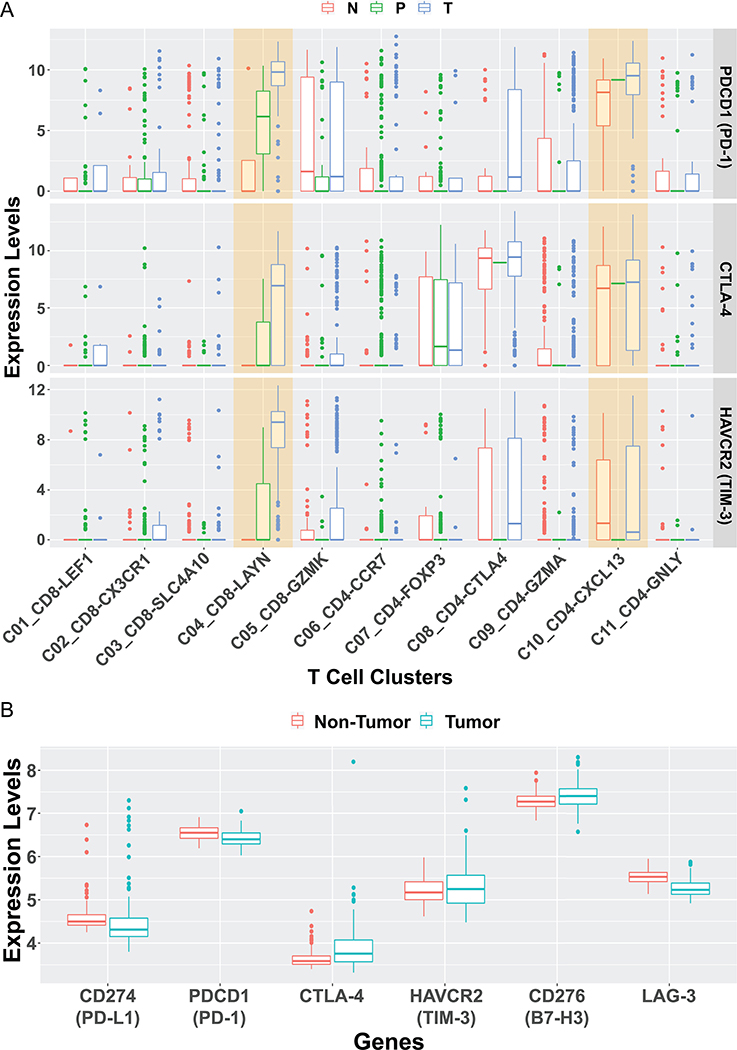
Figure 1. The boxplot of selected immune-inhibitory genes.
A). We re-analyzed the single-cell RNA-seq data published by Zheng et al.(37) The immune-suppressive genes, such as PDCD1 (encoding PD-L1), CTLA4, and HAVCR2 (encoding TIM-3) were compared among various T cell clusters and different tissues. Their expression levels were higher in some T cell clusters from tumor tissue than adjacent normal liver tissue or peripheral blood, with similar levels detected in other clusters. As described,(37) all sequenced T cells were isolated from six treatment-naïve HCC patients and clustered into 11 subpopulations. N: adjacent normal liver tissue; P: peripheral blood; T: tumor tissue. C1: LEF1+, naïve CD8+ T cells; C2: CX3CR1+, effector memory CD8+ T cells; C3: SLC4A10+, MAIT cells; C4: LAYN+, exhausted CD8+ T cells; C5: GZMK+, CD8+ T cells; C6: CCR7+, naïve CD4+ T cells; C7: FOXP3+, peripheral Tregs; C8: CTLA4+, tumor Tregs; C9: GZMA+, CD4+ T cells; C10: CXCL13+, exhausted CD4+ T cells; C11: GNLY+, cytotoxic CD4+ T cells. To generate this plot, TPM values of each gene in all single cells the authors reported, as well as cell description, were downloaded from NCBI Gene Expression Omnibus database (GSE98638). The cells marked as “unknown” were first excluded. Then, the expression for each gene in a single cell was calculated as log(TPM+1) and displayed in boxplots. The calculation and plotting were performed using R.
B). We re-examined six immune-suppressive genes, PD-L1, PD-1, CTLA4, TIM3, B7H3, and LAG3, expressed in another patient cohort, which consisted of 151 surgically paired tumor and non-tumor samples (NCBI Gene Expression Omnibus access code GSE76297) from HCC or CCA patients from Thailand.(38) The microarray data was normalized with Robust Multi-array Average (RMA) and analyzed with GEO2R. No significant difference in the expression of these six genes was detected between the tumor and surrounding non-tumor tissues.