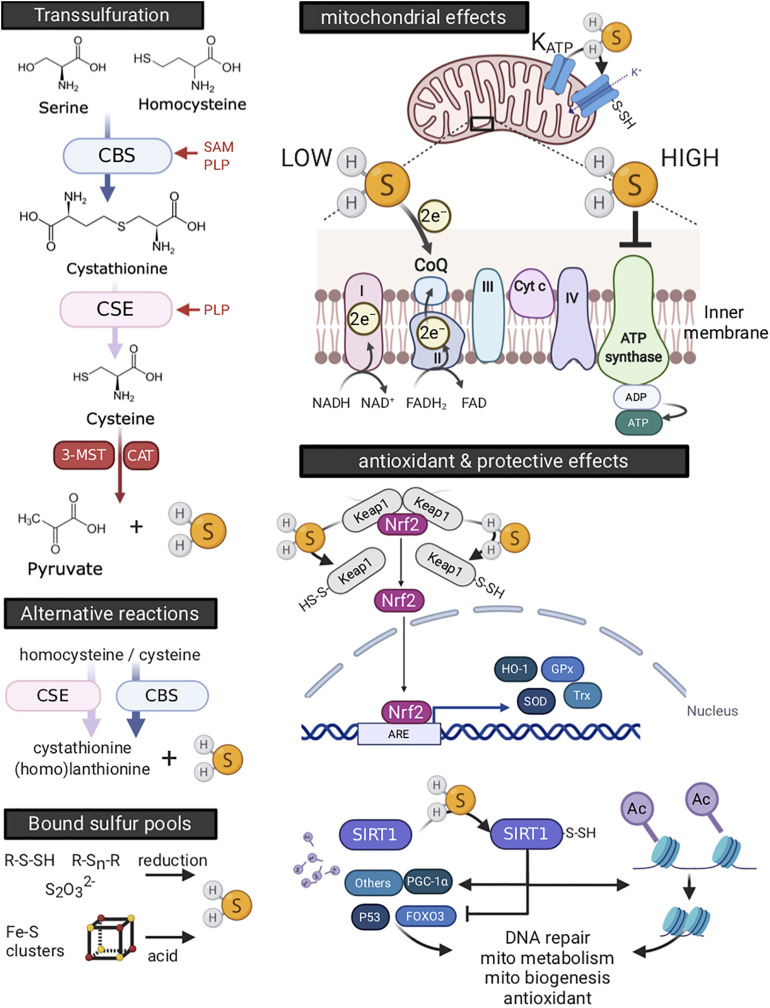
FIGURE 7.
Endogenous production and physiology of hydrogen sulfide (H2S). H2S is enzymatically produced by the trans-sulfuration pathway or by alternative reactions, or derived from compounds binding sulfur (left side panels). H2S producing enzymes differ in various ways, including their dependence on co-factors, cellular, and tissue distribution. Optimal function for both cystathionine β-synthase (CBS) and cystathionine γ-lyase (CSE) is dependent on binding of pyridoxal-5′-phosphate (PLP, or vitamin B6) as a co-factor. In addition, S-adenosyl-L-methionine (SAM), the universal methyl group donor synthesized as an intermediate in the conversion of methionine to homocysteine, serves as a potent allosteric activator of CBS. The cellular localization of CBS and CSE is mainly cytosolic, yet they translocate to mitochondria under stressful conditions (Fu et al., 2012; Teng et al., 2013). 3-mercaptopyruvate sulfur transferase (3-MST) is located in both cytosol and mitochondria, with its activity increasing threefold in mitochondria (Nagahara et al., 1998). Endogenous H2S may also be derived non-enzymatically from “bound sulfur” pools. Upper compounds are sulfane sulfurs, such as thiosulfate (S2O32–), persulfides (R-S-SH) and polysulfides (R-Sn-R), which release H2S in the presence of reducing substances including glutathione (GSH) and cysteine (Ishigami et al., 2009) or by the action of 3-MST (Shibuya et al., 2009). Lower depicted compounds are acid-labile sulfides, which consist mainly of iron-sulfur clusters, of which the cubane-type [4Fe-4S] cluster is most common (Johnson et al., 2005). They are best known for their presence in proteins involved in oxidation-reduction reactions, including those of the electron transport chain in mitochondria. While degradation of iron-sulfur clusters releases H2S, this occurs at strong acidic conditions (pH < 5.4), likely limiting their contribution to endogenous H2S generation even under pathological conditions (Ishigami et al., 2009). H2S affects mitochondria (upper right panel) and produces antioxidant defense (lower right panel). Stimulation of oxygen consumption and ATP production occurs at low H2S concentration and originates from electron donation at coenzyme Q (CoQ) between complexes II and III (Goubern et al., 2007; Módis et al., 2013). At high concentrations, H2S competitively and reversibly inhibits cytochrome c oxidase (complex IV) (Khan et al., 1990; Módis et al., 2014). Moreover, high concentrations of H2S still donate electrons at coenzyme Q, which now reversely flow to complex II, reducing fumarate into succinate (Fu et al., 2012). Moreover, H2S activates the mitochondrial KATP channel (Zhao et al., 2001; Lu et al., 2019) and blocks the Ca2+-mediated opening of the mitochondria permeability transition pore (MPTP) channel, thus supporting maintenance of mitochondrial membrane potential (Yao et al., 2010; Lu et al., 2019; Papu John et al., 2019). In addition, H2S serves as an antioxidant and increases glutathione levels. Further, H2S activates the prominent antioxidant master switch, nuclear factor−erythroid 2−related factor 2 (Nrf2), by sulfhydration of cysteine residues of Kelch-like ECH-associated protein 1 (Keap1) dimer. This breaks Keap1 inhibitory binding to Nrf2 and allows for Nrf2 to translocate to the nucleus (Hourihan et al., 2012; Yang et al., 2012), where it activates expression of antioxidant and cytoprotective genes, such as glutathione peroxidase (Gpx), thioredoxin (Trx), superoxide dismutase (SOD), and heme oxygenase-1 (HO-1). H2S mediated sulfhydration of other proteins confers additional cell protective effects (Mustafa et al., 2009; Zhao et al., 2014). For example, H2S activates the histone deacetylase sirtuin-1 (SIRT1) through its sulfhydration, leading to deacetylation of histones and condensation of chromatin, affecting an array of transcription and nuclear receptors. SIRT1 activation thus influences abundant cellular functions, including stimulation of mitochondrial metabolism and biogenesis, antioxidant pathways, DNA repair and inflammation (Du et al., 2018). ARE, antioxidant response element; Cyt C, cytochrome c, FOXO3, Forkhead box protein O3; PGC-1α, proliferator-activated receptor gamma coactivator 1-alpha. Created with BioRender.com.