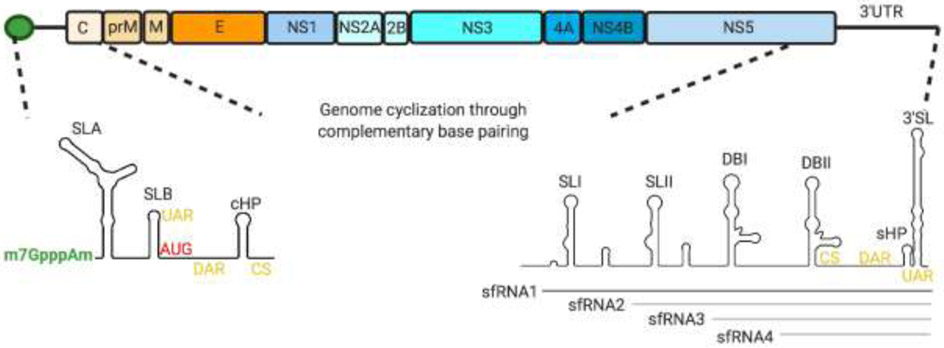
Fig 2:
DENV genome organization. The DENV genomic RNA contains an open reading frame that codes for three structural (C (capsid), prM/M (membrane) and E (envelope)) proteins and seven non structural (NS1, NS2A, NS2B NS3, NS4A, NS4B, NS5) proteins flanked by 5’ and 3’ untranslated regions. The RNA is capped at the 5’ end but lacks a 3’ poly A tail. The 5’UTR is organized into two defined stem loops, SLA which recruits NS5 and SLB. The capsid-coding region hairpin (cHP) downstream of the start codon stalls the PIC at the AUG triplet for translation initiation. The 3’ UTR is organized into stem loops SLI, SLII, dumbbell structures DBI, DBII, a small hairpin (sHP) and a terminal 3’ stem loop (3’SL). There are at least three conserved RNA sequences in the UTRs that mediate genome cyclization through complementary base pairing: 5’-3’ sequence upstream of the AUG region (5’-3’UAR), 5’-3’ sequence downstream of the AUG region (5’-3, DAR) and 5’-3’ cyclization sequence (5’-3’CS) (shown in yellow). The 3’UTR folds to form nuclease resistant structures that cannot be degraded by the host nuclease XrnI, resulting in accumulation of subgenomic flaviviral RNAs (sfRNAs). Four different sfRNA isoforms (sfRNA1-4) can be generated by XrnI stalling at different stem loops on the 3’UTR. sfRNA1 (shown in dark grey) is predominantly formed in human cells while sfRNAs 2-4 (shown in light grey) accumulate in mosquito-adapted variants.