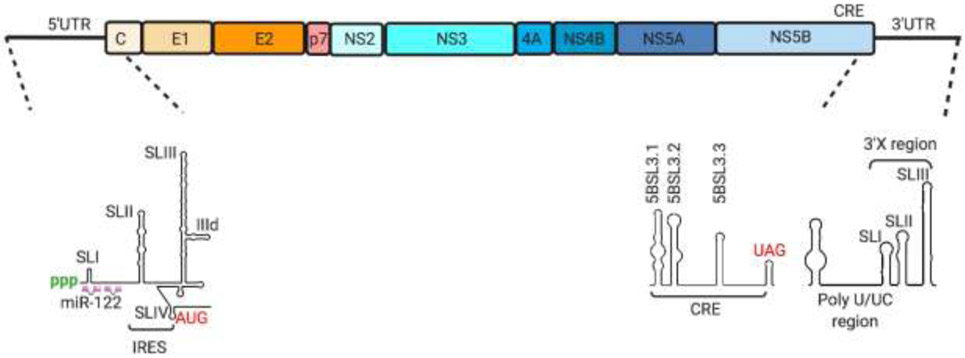
Fig 3.
HCV genome organization. The HCV genomic RNA contains an open reading frame that codes for three structural proteins, core (C) and envelope (E1, E2) and seven non-structural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, NS5B) flanked by 5’ and 3’ untranslated regions. The 5’UTR is organized into stem loops I-IV and harbors the internal ribosome entry site as well as binding sites for miR-122. The 3’UTR is organized into the variable region, the poly U/UC region and the highly conserved 3’X tail, composed of stem loops I-III. The NS5B coding region contains a cis-acting replication element (CRE) at its 3’ end just prior to the 3’UTR. The CRE is organized into three stem loop structures (5BSL3.1, 3.2 and 3.3) which are involved in dynamic interactions with the 5’UTR, 3’UTR and other upstream sequences in the NS5B coding region to regulate translation and replication of the HCV RNA.