Figure 4. Methods for studying eRNAs.
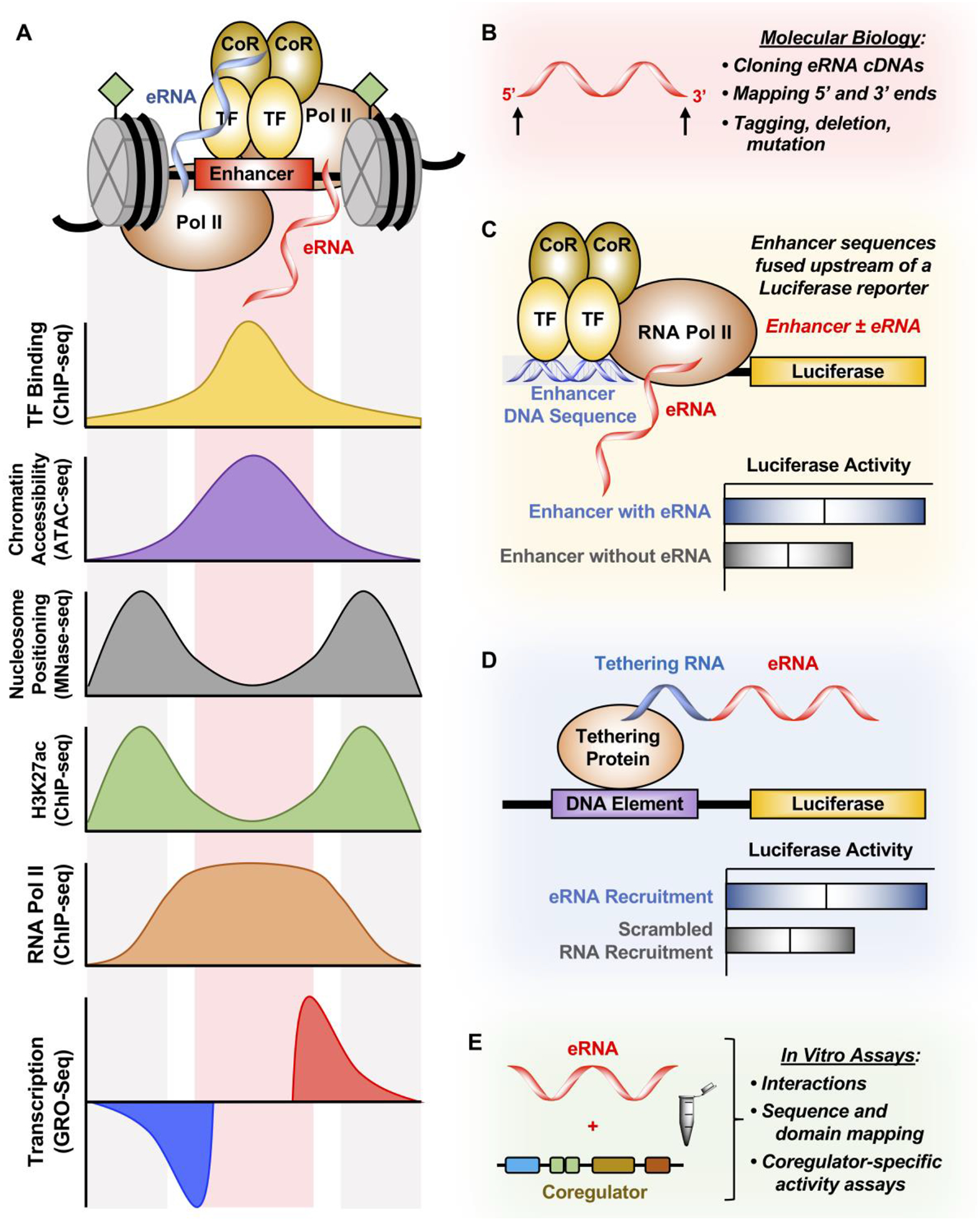
(A) Enhancers are characterized by TF binding (determined by ChIP-seq), increased chromatin accessibility (determined by ATAC-seq), depletion of nucleosomes at TF binding (determined by MNase-seq), enrichment of H3K27ac and RNA Pol II (determined by ChIP-seq), and finally enhancer transcription (determined by GRO-seq). These features are used to derive enhancer region and eRNA ghosts, resulting in a lack of cDNAs derived from eRNAs.
(B) 5’ and 3’ ends of eRNAs can be validated using RACE in order to clone eRNAs into cDNAs to study them by tagging, deleting, and mutating these transcripts.
(C) Enhancer DNA containing or excluding the eRNA sequence derived from genomic sequencing experiments can be cloned upstream of a luciferase reporter to determine the enhancing effect of including the eRNA sequence with the enhancer. However, this technique cannot distinguish between the act of enhancer transcription and the eRNA transcript itself in enhancing luciferase gene transcription.
(D) eRNA can also be tethered to the luciferase reporter construct by using the BoxB-λN or the CRISPR/dCas9 system. This method directly tests whether eRNAs have a function in regulating luciferase gene transcription.
(E) In vitro transcribed eRNAs can be tested for transcription factor binding or coregulator activation in in vitro assays. Additionally, mapping of both the eRNA and the interacting protein can be conducted to further understand how eRNA affects coregulator-specific activity.
