Extended Data Fig. 3. Multi-body refinement of RSCSAR bound to the nucleosome in the ADP-BeF3 state.
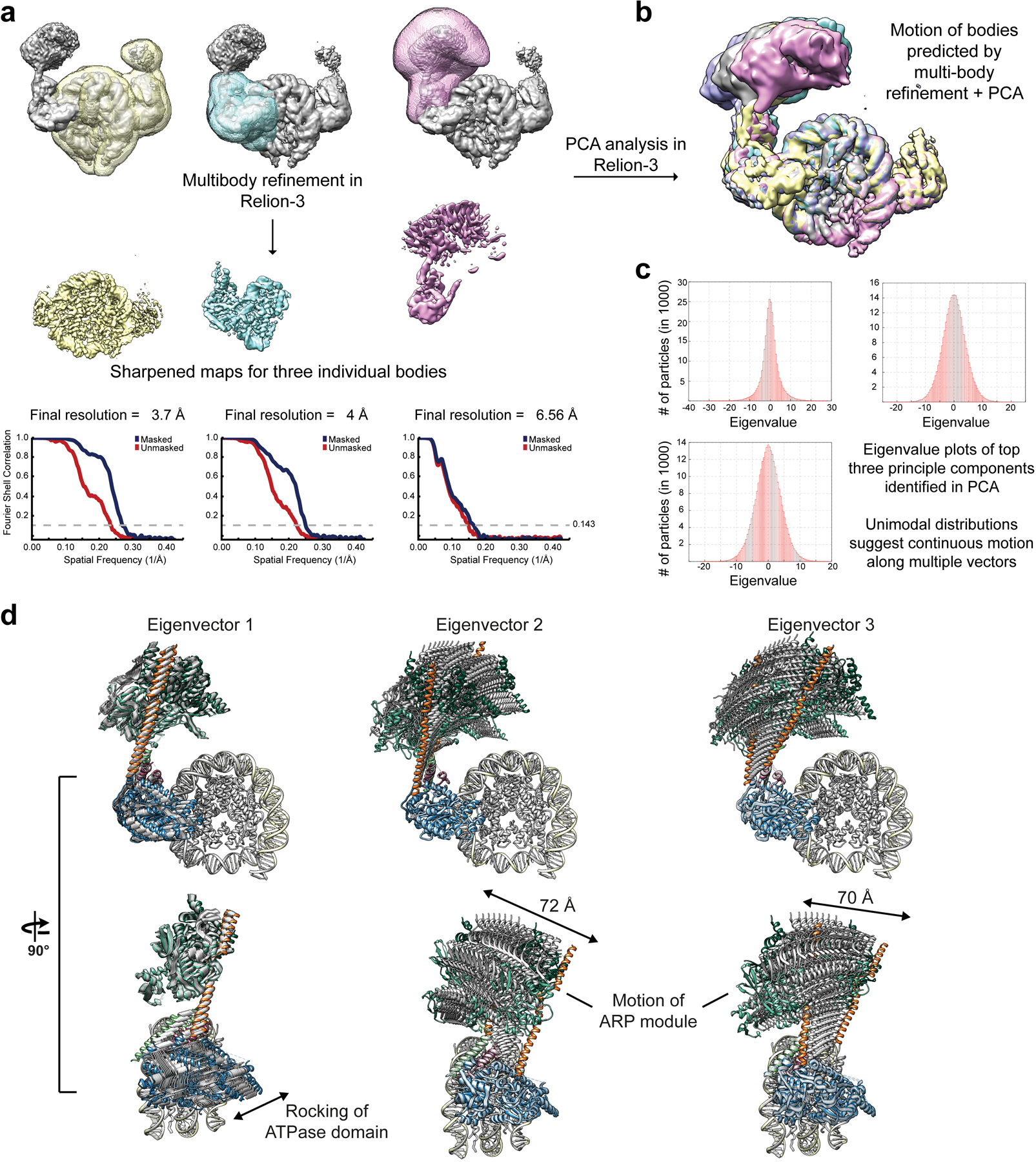
a, The consensus refinement of RSCSAR-nucleosome from Extended Data Fig. 2b was used to generate 3 masks and as a starting model for multi-body refinement. b, PCA was performed in Relion-3 and used to generate maps that show the conformational heterogeneity present in our data set. Several of these maps are shown overlaid. c, Eigenvalue plots for the top three eigenvectors. d, The maps generated in panel (b) were used to build PDB models. 10 models for each eigenvector were generated and are shown superimposed. The main sources of heterogeneity in our data set are a rocking of the ATPase domain (eigenvector 1) and rocking of the ARP module along two different axes (eigenvector 2 and 3).
