Extended Data Fig. 4. Model building of RSCSAR-nucleosome (ADP-BeF3) and comparison with Snf2-nucleosome (ADP-BeF3).
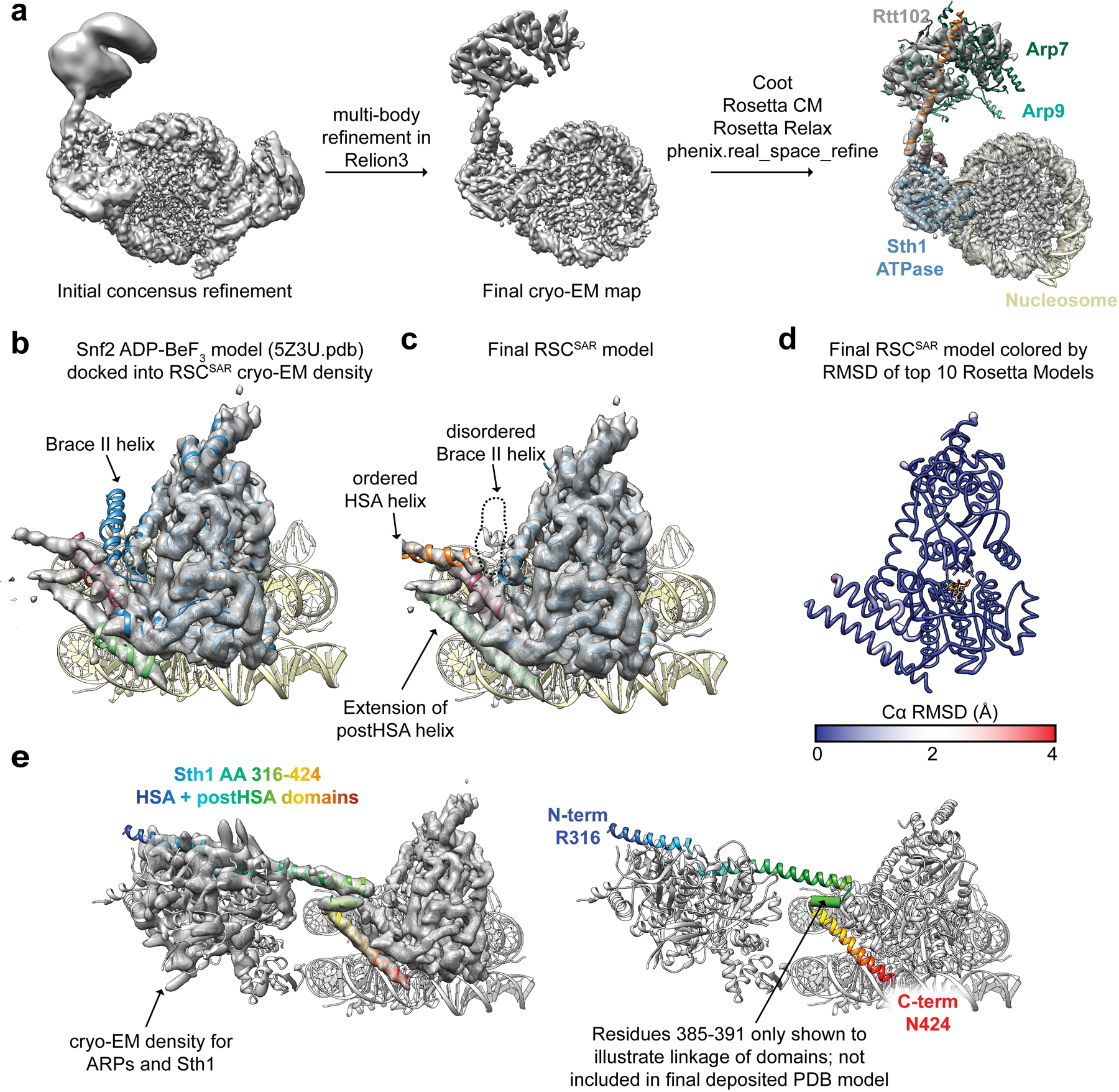
a, Model building schematic for RSCSAR-nucleosome (ADP-BeF3). Snf2-nucleosome (PDB 5Z3U) and the T. thermophilus Snf2 crystal structure (PDB 5HZR) were used as a starting point for model building and refinement using Coot, Rosetta, and phenix.real_space_refine. The ARP module built in Extended Data Fig.1 was rigid-body docked into the map, yielding a near-complete model for all components in our sample. b, PDB 5Z3U is shown docked into our cryo-EM map. c, The same view as in panel (b) but showing Sth1-nucleosome (ADP-BeF3). d, The top Rosetta model is shown with its ribbon thickness and color indicating the RMSD among the top 10 models. e, The RSCSAR-nucleosome structure is shown with the HSA and postHSA domains of Sth1 colored in a rainbow from N to C terminus. Residues 385–391 are shown as a cylinder to signal the linkage between the two domains and indicate that the density is compatible with an alpha helix, although this region is not included in the deposited.
