TO THE EDITOR
Atopic dermatitis (AD) is a common inflammatory skin disease which manifests as itchy, red patches appearing early in life (Abramovitz et al.,2005;Margolis et al.,2014b). Previous studies have associated TSLP, a cytokine which promotes the differentiation of type 2 helper T cells, and specifically the single nucleotide polymorphism (SNP) rs1898671 (TSLP:g.110408002C>T), with the prevalence and persistence of AD (Gao et al.,2012;Margolis et al.,2014a). TSLP exerts its action via a heterodimeric receptor complex composed of IL-7Rα (encoded by the gene IL7R) and TSLPR (encoded by CRLF2) (Fornassa et al.,2015).
Study of TSLP and its receptor is important for understanding the genetic architecture of AD and identifying potential therapeutic targets (Margolis et al.,2014a). Prior studies of TSLP and IL7R have been limited to the examination of tagging SNPs (Gao et al.,2012; Hoffjan et al.,2009). In this report, we fully sequenced the TSLP and IL7R genes in a large longitudinal cohort of white and African American individuals with AD to gain more complete insight into their association with AD persistence.
Data were obtained from the Pediatric Elective Eczema Registry (PEER), a longitudinal cohort of children with mild-to-moderate AD, for whom disease control was assessed via self-report every 6 months for up to 10 years. Disease was considered persistent if the participant did not report “complete disease control” over the past six months and did not use prescription cream. Multiple measurements of this outcome were recorded over time for each participant. Next generation sequencing (NGS) of TSLP and IL7R was conducted on 739 individuals in PEER. Only the TSLP and IL7R genes were targeted for sequencing. Association between the outcome measure and individual SNPs was evaluated using generalized estimating equations (Berna et al.,2020). Full methods are described in the Supplement. Written, informed consent was obtained from all enrolled children. This study was approved by the Institutional Review Board of the University of Pennsylvania.
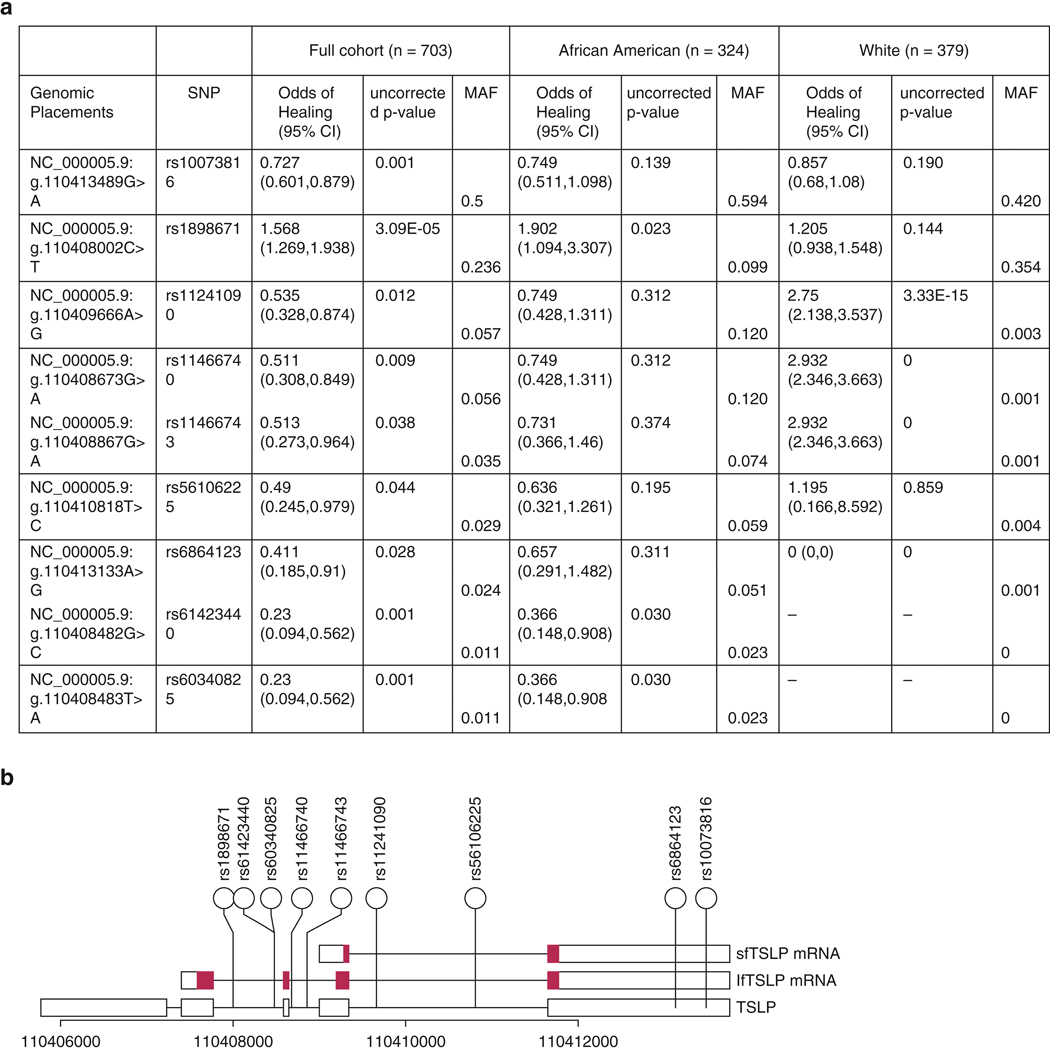
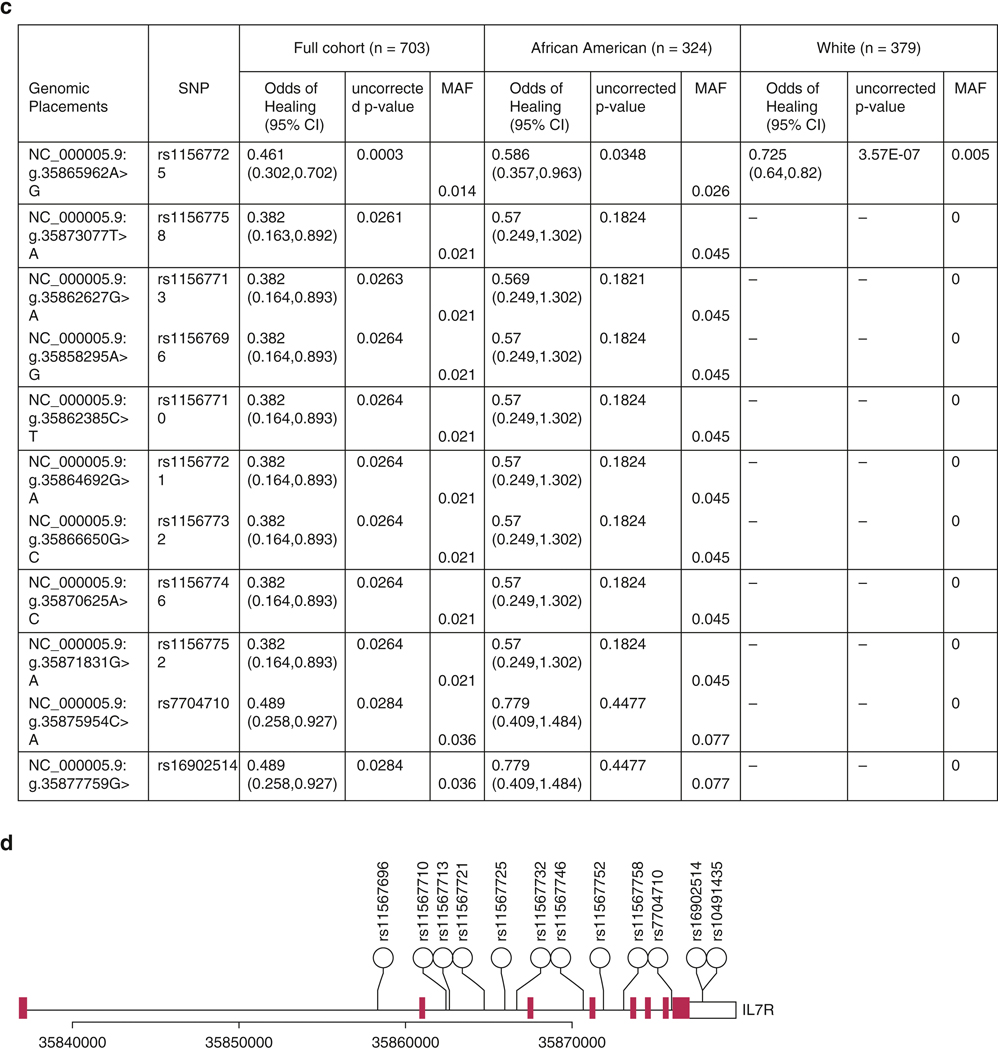
703 individuals described themselves as either white (n=379) or African American (n=324). Participant demographics are presented in Table 1. For the TSLP gene, 161 distinct SNPs were identified, 30 with MAF≥1%. Within the IL7R gene, 428 distinct SNPs were identified, 144 with MAF≥1%. Figure 1 presents associations of TSLP and IL7R SNPs with the persistence of AD over time in the full cohort and separately by race. The rs10073816 SNP (TSLP:g.110413489G>A) retained statistical significance after multiple testing correction in the full cohort (p=0.001). rs61423440 (TSLP:g.110408482G>C) and rs60340825 (TSLP:g.110408483T>A) also retained significance, after correction, in the full cohort (p=0.001,p=0.001,respectively). Within IL7R, the rs11567725 SNP (IL7R:g.35865962A>G) was associated with increased AD persistence in the full cohort, after correction (p=0.0003).
Table 1.
Participant Demographics
| Full cohort, n (%) | African American, n (%) | White, n (%) | p value (African American vs. White) | |
|---|---|---|---|---|
| Number | 703 | 324 | 379 | |
| Age of AD onset, mean (SD) | 1.98 (2.74) | 2.14 (2.85) | 1.84 (2.65) | 0.162 |
| Sex: male, n (%) | 332 (47.23) | 136 (41.98) | 196 (51.72) | 0.012 |
| Asthma, n (%) | 382 (54.34) | 182 (56.17) | 200 (52.77) | 0.408 |
| Seasonal Allergies, n (%) | 494 (70.27) | 228 (70.37) | 266 (70.18) | ~1 |
| Observation time in months, mean (95% CI) | 117.3 (115.8,118.8) | 116.9 (115.2,118.8) | 117.7 (115.8,119.4) | 0.549 |
Figure 1. Evaluation of TSLP and IL7R SNPs.
(a) Association of TSLP SNPs with outcome, ordered by number of allele occurrences. Only variants with raw p values in the pooled analysis ≤ 0.05 and MAF ≥ 1% are reported. Presented p values are all raw. Bolded SNPs have p-values which satisfy Bonferroni correction in the pooled analysis (adjusted p = 0.00167). Genomic placements are from sequence GRCh37.p13 chr 5. (b) Schematic representation of human TSLP based on Fornassa et al. (hg 19), with locations of SNPs from the table above. Blue regions are coding. (c) Association of IL7R SNPs with AD persistence. Only variants with raw p values in the pooled analysis ≤ 0.05 and MAF ≥ 1% are reported. Presented p values are all raw. Bolded SNPs have p-values which satisfy Bonferroni correction in the pooled analysis (adjusted p = 0.00035). Genomic placements are from sequence GRCh37.p13 chr 5. (d) Schematic representation of human IL7R based on the NCBI gene database (hg 19), with locations of SNPs from the table above. Blue regions are coding.
Rs10073816 was associated with more persistent AD and is not in strong linkage disequilibrium (R2>0.6) with other TSLP SNPs with MAF≥1%, with the exception of rs2289277 (TSLP:g.110409067C>G), R2=0.61, an SNP not associated with AD persistence (Supplementary Figure 1). eQTL analysis provided in GTEx demonstrates that rs10073816 is negatively associated with TSLP mRNA expression in sun-exposed skin, non-sun-exposed skin, and cultured fibroblasts (p=8.6e-39,1.1e-35,5.3e-18,respectively).
TSLP variant rs10073816 was more strongly associated with AD persistence when controlling for IL7R variant rs11567725 in both the full cohort (OR,0.646;95%CI,0.525–0.796;p=3.961e-5) and African American population (OR,0.609;95%CI,0.409–0.908;p=0.0148).
Our study demonstrates considerable variation in TSLP and IL7R; however, little is associated with AD persistence. We identify variants previously unassociated, to our knowledge, with AD, including rs61423440 and rs60340825 (MAF<5%), associated with a 4-fold increase in AD persistence, and rs10073816 (MAF=50.1%), associated with a 1.4 fold increase in AD persistence. The rs10073816 association was only observed in the full cohort, likely because this population is larger than the African American or white populations alone.
Our rs10073816, rs61423440, and rs60340825 associations have not, to our knowledge, been observed in other studies, even though a number of analyses of TSLP have been conducted (Gao et.al.,2012;Lou et al.,2019;Margolis et.al.,2014a;Ziegler,2012). The primary difference between our study and previous reports is that we used NGS to identify variants.
Identified TSLP variants are located in non-coding regions, suggesting that TSLP variation in AD is linked to altered protein quantity, not structure. The TSLP gene is transcribed into two proteins—a long isoform which is proinflammatory and a short isoform which is anti-inflammatory (Fornassa et al.,2015). The fact that rs10073816 is associated with decreased TSLP mRNA production and not in LD with any variants equally associated with AD persistence suggests that its effect on AD persistence may be causal. Since AD is thought to be an inflammatory disorder and rs10073816 is associated with decreased production of TSLP, it is likely that rs10073816 is associated with decreased production of TSLP’s short isoform.
It is reasonable to think that IL7R, a component of the TSLP receptor, could be associated with AD persistence. One SNP, rs11567725 (MAF~1%), was associated with increased AD persistence. Our data imply that further study of IL7R may not uncover variants, at a clinically important frequency, associated with AD. The fact that variants in IL7R do modulate AD persistence suggests that this receptor protein may still be a valuable therapeutic target in AD.
This report’s primary limitation is that SNPs in the TSLP and IL7R genes with MAF<1% were not analyzed. These SNPs may be associated with variation in AD severity and important in understanding the etiology of genetically distinct forms of AD. Although our sample size is large for a full-gene sequencing study (>700 individuals), these rare variants were difficult to analyze due to limited statistical power. However, our primary interest was the identification of clinically-actionable mutations; rare variants are less useful in clinical practice. We therefore a priori only evaluated common SNPs.
In summary, we have conducted NGS of the TSLP and IL7R genes and identified that TSLP variants rs10073816, rs61423440, and rs60340825 are associated with more persistent AD. The rs10073816 variant is very common in the population, associated with decreased TSLP mRNA production, and not in strong LD with any variants equally likely to affect AD persistence. IL7R had only one mutation, rs11567725, associated with AD persistence. IL7R variants may modulate the effect of TSLP variants, for the association of rs10073816 with AD persistence is strengthened when controlling for rs11567725. This study strongly supports additional investigation of the TSLP pathway in AD, encourages further study of therapeutics targeting TSLP and IL7R, and promotes the case for including TSLP in genetic risk models for AD persistence.
Supplementary Material
FUNDING AND ACKNOWLEDGMENTS
This project has been funded in whole or in part by R01-AR060962 and R01-AR070873 from NIAMS (PI: David Margolis). The PEER study is funded by Valeant Pharmaceuticals (Pi: David Margolis). The sponsors did not have a role in the design of the study, the collection, analysis, or interpretation of the data, the preparation of this report, or the decision to submit this report for publication.
Abbreviations:
- AD
atopic dermatitis
- CI
confidence interval
- FLG
filaggrin
- LoF
loss of function
- TSLP
thymic stromal lymphopoietin
- PEER
Pediatric Eczema Elective Registry
- MPS
massively parallel sequencing
- MAF
minor allele frequency
- GEE
generalized estimating equations
Footnotes
DATA AVAILABILITY
Datasets related to this article can be found within the online data repository hosted by Mendelay at DOI: 10.17632/sgnkksb36x.1 under the title: “Datasets for ‘Thymic Stromal Lymphopoietin and IL7R Variants are Associated with Persistent Atopic Dermatitis.’”
CONFLICT OF INTEREST
David Margolis is a consultant for Pfizer, Leo, and Sanofi with respect to studies of atopic dermatitis and serves on an advisory board for the National Eczema Association. Joy Wan receives research funding from Pfizer, Inc for work unrelated to this study. No other authors state financial conflicts of interest with respect to this investigation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abramovitz W. Atopic dermatitis. J Am Acad Dermatol. 2005;53(1)(suppl 1):S86–S93. [DOI] [PubMed] [Google Scholar]
- Abuabara K, Margolis DJ. Do children really outgrow their eczema, or is there more than one eczema? J Allergy Clin Immunol. 2013; 132(5):1139–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna R, Mitra N, Hoffstad O, Wan J, Margolis DJ. Identifying phenotypes of atopic dermatitis in a longitudinal US cohort using unbiased statistical clustering. J Invest Dermatol. 2020;140(2):477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornassa G, Tsilingiri K, Caprioli F, Botti F, Mapelli M, Meller S, et al. Dichotomy of short and long thymic stromal lymphopoietin isoforms in inflammatory disorders of the bowel and skin. J Allergy Clin Immunol. 2015;136(2):413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewcz M, et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2012;125(6):1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffjan S, Beygo J, Akkad DA, Parwez Q, Petrasch-Parwez E, Epplen JT. Analysis of variation in the IL7RA and IL2RA genes in atopic dermatitis. J Dermatol Sci. 2009; 55(2):138–140. [DOI] [PubMed] [Google Scholar]
- Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003; 361(9352):151–160. [DOI] [PubMed] [Google Scholar]
- Lou C, Mitra N, Wubbenhorst B, D’Andrea K, Hoffstad O, Kim BS, et al. Association between fine mapping thymic stromal lymphopoietin and atopic dermatitis onset and persistence. Ann Allergy Asthma Immunol. 2019;123(6):595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Kim B, Apter AJ, Gupta J, Hoffstad O, Papadopoulos M, et al. Thymic stromal lymphopoietin variation, filaggrin loss of function, and the persistence of atopic dermatitis. JAMA Dermatology. 2014; 150(3):254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis JS, Abuabara K, Bilker W, Hoffstad O, Margolis DJ. Persistence of mild to moderate atopic dermatitis. JAMA Dermatol. 2014b; 150(6):593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler SF. Thymic stromal lymphopoietin (TSLP) and allergic disease. J Allergy Clin Immunol. 2012; 130(4):845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.