Extended Data Fig. 9: Intrinsic cyclizability along nucleosomes.
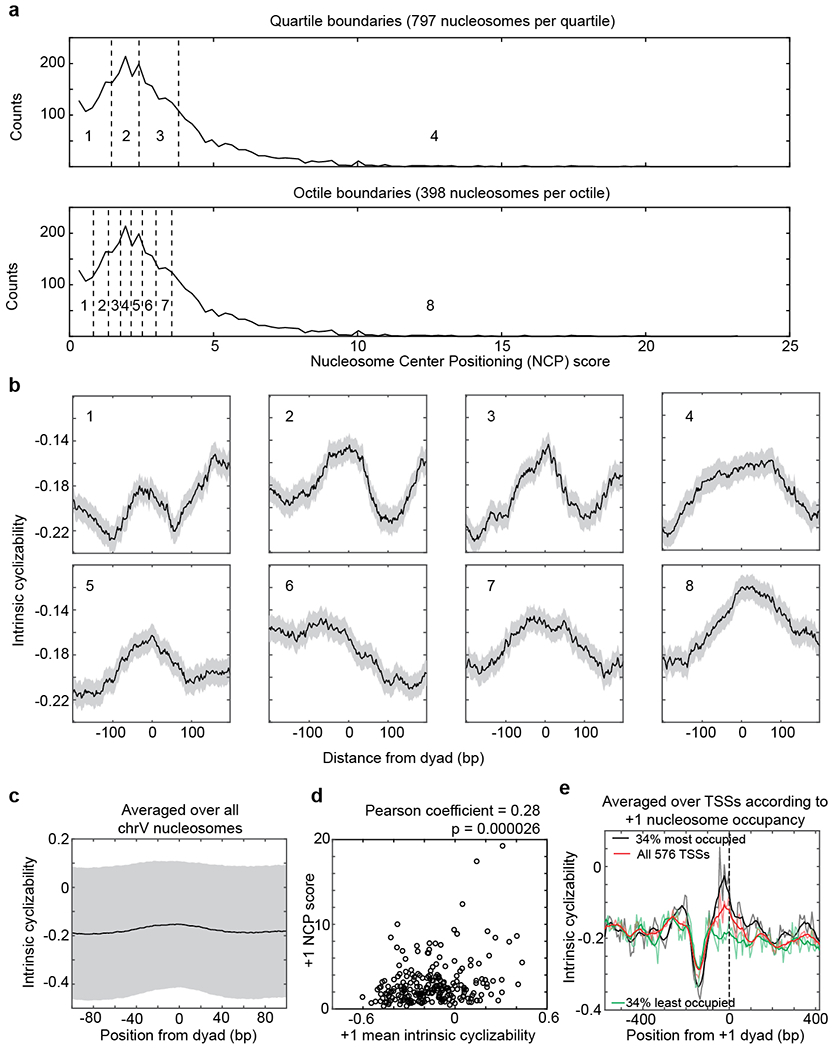
a, Distribution of Nucleosome Center Positioning (NCP) scores32 of all 3,192 S. cerevisiae chromosome V nucleosomes. Quartile and octile boundaries of the distribution are shown in dashed lines and numbered (1 through 4 for quartiles and 1 through 8 for octiles). b, Mean intrinsic cyclizability of DNA as a function of position from the dyads of nucleosomes along yeast chromosome V, averaged over nucleosomes in each octile indicated in panel a. Error extents (shaded background) are s.e.m. c, Mean intrinsic cyclizability as a function of position, averaged over all 3,192 S. cerevisiae chromosome V nucleosomes (solid line). Height of the shaded region is the standard deviation of measurements. d, Scatter plot of the NCP scores of the 227 +1 nucleosomes of the 227 genes identified along chromosome V vs the mean intrinsic cyclizabilities of DNA along the 147 bp that span these nucleosomes. Intrinsic cyclizability values were obtained by performing loop-seq on the ChrV Library. Pearson’s r = 0.28. 95% CI = [0.15, 0.39]. p = 2.6E-5 (t-test, two-sided). e, Plot of intrinsic cyclizability as a function of position along all the 576 genes in the Tiling Library (red), and among 34% of these genes that had the highest (black) or lowest (green) NCP score value of the gene’s +1 nucleosome. Plots were obtained in a manner identical to that in Fig. 2b. Intrinsic cyclizability on either side of the dyad of +1 nucleosomes of genes that have high +1 nucleosome NCP score (black) is asymmetric, being higher on the TSS proximal (i.e. “left”) side of the dyad.
