Figure 3. DNA mechanics impacts chromosome-wide nucleosome organization.
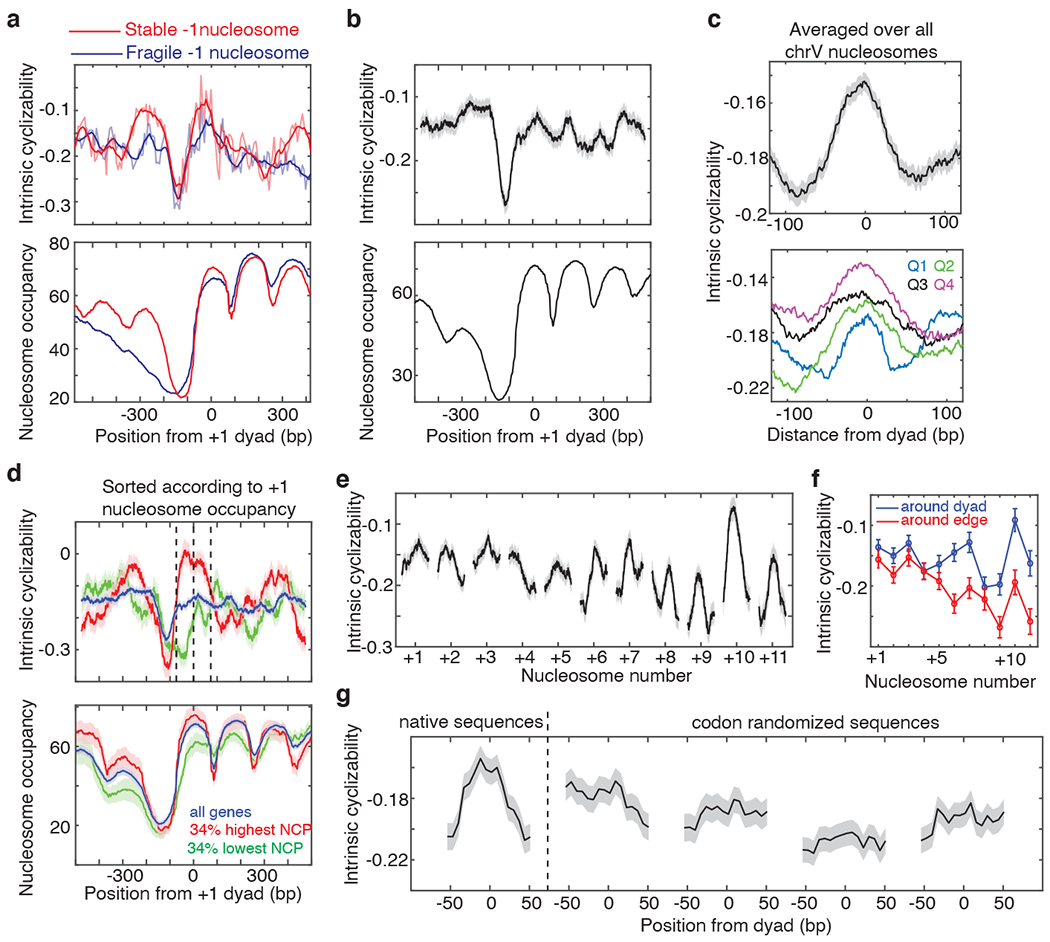
Mean intrinsic cyclizability and nucleosome occupancy vs position from the dyad of the +1 nucleosome, (a) averaged over 185 and 345 genes in the Tiling Library which possess stable and fragile −1 nucleosomes respectively24 (plotted as in Fig 2b), and (b) averaged over all 227 identified genes along S. cerevisiae chromosome V (Supplementary Note 13). Grey background: s.e.m. (c) Intrinsic cyclizability vs position from the dyad, averaged over all 3,192 nucleosomes along chromosome V (top), or over nucleosomes sorted into quartiles based on reported NCP scores32 (bottom). See also Extended Data Fig. 8a–b. Solid lines: mean, grey background: s.e.m. (d) Intrinsic cyclizability and nucleosome occupancy (solid lines) vs position along all chromosome V genes (blue), and among 34% of genes with the highest (red) and lowest (green) +1 nucleosome NCP scores32. Plots were obtained as in panel b. Dashed lines: edges and dyad of the +1 nucleosome. Shaded backgrounds: s.e.m. (e) Intrinsic cyclizability around nucleosomal dyads that lie within the transcribed region of all identified 227 genes along chromosome V in S. cerevisiae. Solid lines: mean, grey background: s.e.m. See supplementary Note 13 for N values. (f) Mean and s.e.m. of intrinsic cyclizability in a 50 bp window around the dyads (blue) and edges (red, from position −73 till −56 and from +56 till +73) of gene-body nucleosomes. N values are the same as in panel e. (g) Intrinsic cyclizability of the native sequences around the dyads of the 500 +7 nucleosomes represented in Library L (supplementary note 14), and along four sets of codon-altered sequences generated by randomly selecting synonymous codons while considering (the first two) or not considering (the next two) the natural codon-usage frequency. Solid line: mean, smoothened over a 7-fragment rolling window, grey background: s.e.m.
