Extended Data Fig. 1. Activation loop mutagenesis of Mec1.
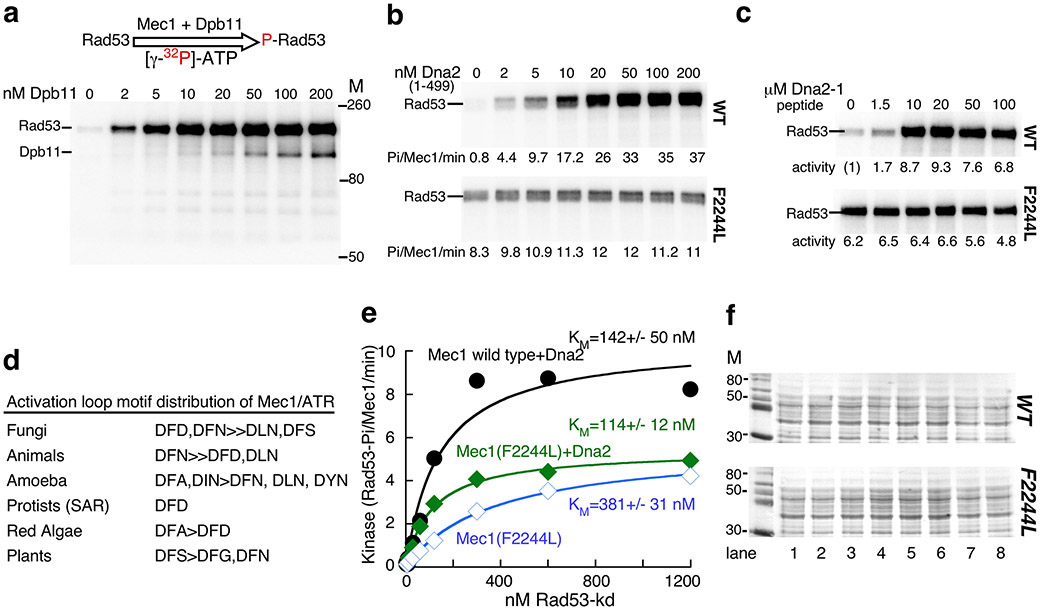
(a) Representative complete gel of Mec1 kinase assay, described in Fig. 2. The gel shown is for the experiment in Fig. 2a, WT.
(b) Kinase activity of Mec1 and Mec1(F2244L) as a function of Dna2(1-499). Phosphorylation rates are given below the gel.
(c) Kinase activity of Mec1 and Mec1(F2244L) as a function of Dna2-1 peptide: HHDFTQDEDGPMEEVIWKYSPLQRDMSDKT. Fold stimulation compared to wild-type Mec1 without activator is given.
(d) Phylogenetic analysis of the activation loop 2243DFD2245 motif. 640 eukaryotic Mec1/ATR sequences were aligned with MSAProbs (https://toolkit.tuebingen.mpg.de/), filtered to a set of 95 sequences that showed less than 50% sequence identity, and the motif distribution recorded.
(e) Titration of Rad53 into the Mec1 assay. Standard assays with 3 nM Mec1 and 5 nM Dna2(1-499) activator, or with 3 nM Mec1(F2244L) with or without 5 nM Dna2(1-499) activator were carried out at increasing concentrations of GST-Rad53-kd. Activities are expressed as Rad53 phosphates per Mec1 (monomer) per minute, and the data were modeled to the Michaelis-Menten equation.
(f) Ponceau staining of the extracts used for the Western blots in Fig. 2e.