Extended Data Fig. 10 |. Potential chemical modulation of radial spokes.
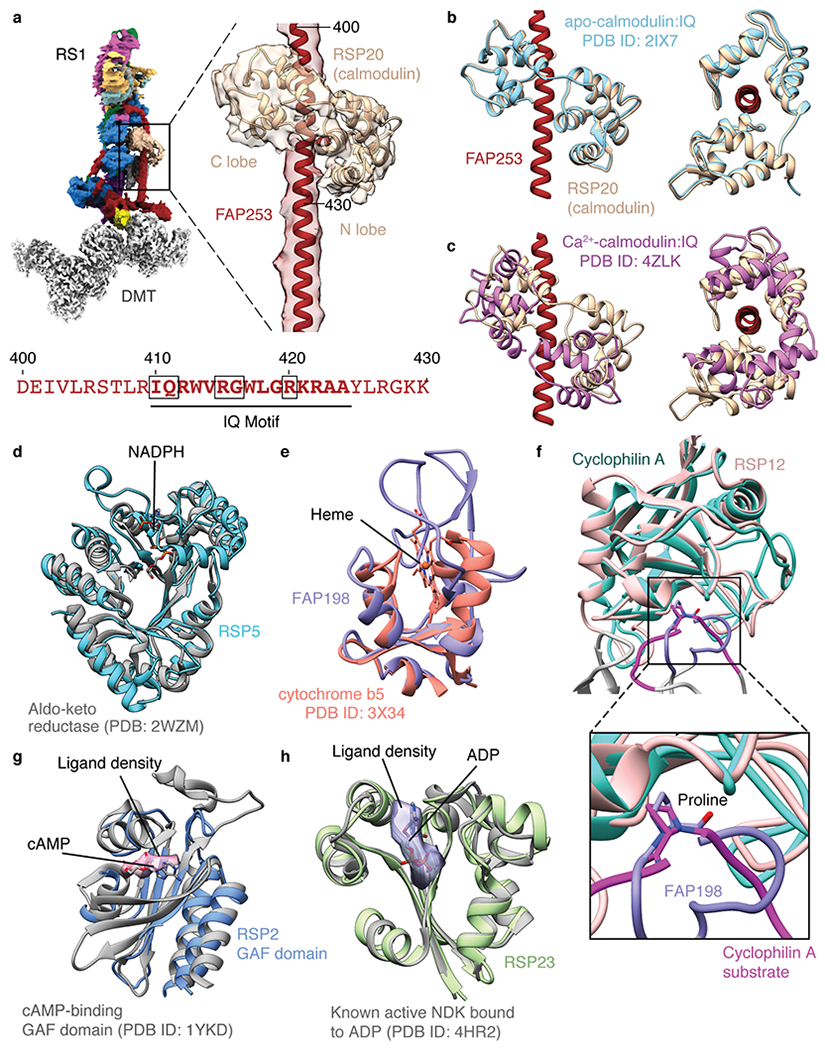
a, Calmodulin binds the IQ motif of FAP253 at the base of RS1. Below, sequence of FAP253 residues 400-430 showing the presence of an IQ motif (emboldened with motif-defining residues boxed). b, Structural comparison of calmodulin bound to FAP253 with apo-calmodulin bound to an IQ motif from myosin V (PDB 2IX7)42. c, Structural comparison of calmodulin bound to FAP253 with Ca2+-calmodulin bound to an IQ motif from myosin 5a (PDB 4ZLK)42. d, The structure of RSP5 resembles an NADPH-dependent aldo-keto reductase domain (PDB 2WZM)76. However, the NADPH binding site of RSP5 is absent and filled by two loops (residues 393-414 and 468-484 of RSP5). e, The N-terminal domain of FAP198 closely resembles heme-binding cytochrome b5 (PDB 3X34)77. However, no heme is observed bound to FAP198, and the putative heme-binding site is occluded by a loop of FAP198 (residues 89-95). f, RSP12 structurally resembles cylophilin-type peptidyl-prolyl cis-trans isomerase (PDB 1AK4)78. The putative substrate-binding site of RSP12 is occupied by a loop of FAP198 (residues 96-105), which positions a proline (P99) in the active site. g, Atomic model of the GAF domain from RSP2 superposed with the model of a cAMP-bound GAF domain (PDB 1YKD)79. Unexplained density in the RSP2 GAF domain (pink, contoured at 0.01) is observed in the cAMP binding pocket, but the resolution is insufficient to assign it as a cyclic nucleotide. The cAMP ligand from PDB 1YKD is shown for comparison. h, Atomic model of RSP23 superposed with an active, ADP-bound nucleoside diphosphate kinase (NDK; PDB 4HR2). Many of the active site residues are conserved. Potential density for a bound nucleotide to RSP23 is observed in the on-doublet map of RS1 (purple, contoured at 0.017) but not in the isolated RS1 map. The ADP ligand from PDB 4HR2 is shown for comparison.
