Abstract
PNPLA6-related disorders include several phenotypes, such as Boucher–Neuhäuser syndrome, Gordon Holmes syndrome, spastic paraplegia, photoreceptor degeneration, Oliver-McFarlane syndrome and Laurence-Moon syndrome. In this study, detailed clinical evaluations and genetic testing were performed in five (4 Chinese and 1 Caucasian/Chinese) syndromic retinal dystrophy patients. Genotype-phenotype correlations were analyzed based on review of the literatures of previously published PNPLA6-related cases. The mean age of patients and at first visit were 20.8 years (11, 12, 25, 28, 28) and 14.2 years (4, 7, 11, 24, 25), respectively. They all presented with severe chorioretinal dystrophy and profoundly decreased vision. The best corrected visual acuity (BCVA) ranged from 20/200 to 20/2000. Systemic manifestations included cerebellar ataxia, hypogonadotropic hypogonadism and hair anomalies. Six novel and three reported pathogenic variants in PNPLA6 (NM_001166111) were identified. The genotypes of the five cases are: c.3134C>T (p.Ser1045Leu) and c.3846+1G>A, c.3547C>T (p.Arg1183Trp) and c.1841+3A>G, c.3436G>A (p.Ala1146Thr) and c.2212–10A>G, c.3436G>A (p.Ala1146Thr) and c.2266C>T (p.Gln756*), c.1238_1239insC (p.Leu414Serfs*28) and c.3130A>G (p.Thr1044Ala). RT-PCR confirmed that the splicing variants indeed led to abnormal splicing. Missense variants p.Thr1044Ala, p.Ser1045Leu, p.Ala1146Thr, p.Arg1183Trp and c.3846+1G>A are located in Patatin-like phospholipase (Pat) domain. In conclusion, we report the phenotypes in five patients with PNPLA6 associated syndromic retinal dystrophy with variable systemic involvement and typical choroideremia-like fundus changes. Ocular manifestations may be the first and the only findings for years. All of our patients carried one severe deleterious variant (stop-gain or splicing variant) and one milder variant (missense variant). Retinal involvement was significantly correlated with severe deleterious variants and variants in Pat domain.
Keywords: PNPLA6 gene, syndromic retinal dystrophy, Boucher-Neuhäuser syndrome, Oliver-McFarlane syndrome, Choroideremia-like
1. Introduction
Patatin-like phospholipase domain containing 6 (PNPLA6) is a highly conserved phospholipase also known as neuropathy target esterase (NTE), which deacetylates intracellular phosphatidylcholine to produce glycerophosphocholine(Zaccheo et al., 2004; Sogorb et al., 2016; Glynn, 2003). The PNPLA6 gene maps to chromosome 19p13.2, and five different transcripts have been identified, with the longest transcript, transcript variant 1 (NM_001166111), encoding a protein of 1,375 amino acids. PNPLA6 plays an important role in multiple systems and is speculated to be involved in phosphatidylcholine metabolism, neuronal development, intercellular membrane trafficking, axon maintenance and is the target in organophosphate-induced delayed neuropathy (OPIDN)(Synofzik et al., 2014; Richardson et al., 2013).
PNPLA6 was first linked to disease in 2008(Rainier et al., 2008), and has been identified in patients diagnosed with Boucher–Neuhäuser syndrome, Gordon Holmes syndrome, spastic paraplegia, retinal degeneration, Oliver-McFarlane syndrome and Laurence-Moon syndrome(Synofzik et al., 2014; Hufnagel et al., 2015; Kmoch et al., 2015; Synofzik et al., 2015). PNPLA6-related disorders have variable clinical phenotypes and span a phenotypic continuum characterized by variable combinations of cerebellar ataxia, chorioretinal dystrophy, hypogonadotropic hypogonadism, peripheral neuropathy, hair anomalies, short stature, and intellectual disability. No obvious genotype–phenotype correlation has been proposed in patients with PNPLA6 variants and all the associated diseases are very rare. Overlapped phenotypic features exist among different symptoms and therefore it is challenging to make a diagnosis based on clinical manifestations(Synofzik et al., 2015).
To the best of our knowledge, only about 65 cases with biallelic PNPLA6 variants have been reported worldwide (hgmd.org) (Zheng et al., 2018; D’Amore et al., 2018; Coutelier et al., 2018; Stone et al., 2017; Teive et al., 2018; Hufnagel et al., 2015; Tarnutzer et al., 2015; Kmoch et al., 2015; Koh et al., 2015; Synofzik et al., 2014; Topaloglu et al., 2014; Fogel et al., 2014; Deik et al., 2014; Yoon et al., 2013; Rainier et al., 2008; Patsi et al., 2018; Wiethoff et al., 2017; Langdahl et al., 2017; O’Neil et al., 2019; Salgado et al., 2019; DeNaro et al., 2018; Rainier et al., 2011). Most previous studies on PNPLA6-related disorders were performed in the western population, and only one Chinese patient was available(Zheng et al., 2018). Chorioretinal dystrophy is a common clinical feature shared by Boucher–Neuhäuser syndrome (BNS), Oliver-McFarlane syndrome (OMS) and Laurence-Moon syndrome (LMS). We find that vision impairment is often the initial symptom. However, an awareness of the PNPLA6-related disorders is low among ophthalmologists and the PNPLA6 syndromes are prone to be misdiagnosed because of their rarity and complexity. Frequently, previous reports lacked detailed description of ocular manifestations in PNPLA6-related disorders. In this study, we report the detailed clinical features, especially ocular features, in four Chinese patients and one Caucasian/Chinese patient with biallelic PNPLA6 variants. Additionally, we provide an up to date review of all published PNPLA6-related cases to date.
2. Materials and Methods
2.1. Recruitment of subjects
Participants were enrolled at Peking Union Medical College Hospital (PUMCH), Beijing, China and from the Oregon Health & Science University (OHSU) – Casey Eye Institute, Portland, Oregon, USA. This study was approved by the Institutional Review Board of PUMCH and OHSU and adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained from each participant.
2.2. Clinical evaluations
Detailed medical history and family history were obtained for all affected patients. Full ophthalmological examinations, including best corrected visual acuity (BCVA), intraocular pressure, detailed slit-lamp examination, dilated indirect ophthalmoscopy, fundus photography (Topcon, Tokyo, Japan and Optos Inc., Marlborough, MA, USA), static perimetry (Octopus, Interzeag, Schlieren, Switzerland or Humphrey, Zeiss, Dublin, California, USA) or kinetic perimetry, optical coherence tomography (OCT; Heidelberg HRT II, Heidelberg, Germany), fundus autofluorescence imaging (Heidelberg HRT II, Heidelberg, Germany and Optos Inc., Marlborough, MA, USA), full-field electroretinography (ERG; Roland Consult, Wiesbaden, Germany). Magnetic resonance imaging (MRI) of the brain and hormonal studies were conducted for all patients.
2.3. Genetic testing
Peripheral blood samples were collected, and genomic DNA was extracted from all affected subjects and unaffected available family members with the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. A customized retinal disease gene target capture with next-generation sequencing was performed first in two patients as previously described(Jiang et al., 2015). And multiplex ligation-dependent probe amplification (MLPA) was performed in the same two patients with SALSA multiplex ligation-dependent probe amplification probe mix P366-A2 CHM-RP2-RPGR (Lot A2–0614; MRC-Holland, Amsterdam, the Netherlands) according to the manufacturer’s instructions. The detailed procedure of MLPA was described previously(Zhou et al., 2017). Subsequently, all patients’ DNA underwent whole exome sequencing (WES). Starting with 200ng high quality genomic DNA, libraries were constructed using KAPA HyperPlus Kits (Kapa Biosystems) by following the manufacture’s instruction. The post-PCR library was then used for exome capture using the Fulgent WESPlus panel, an enhanced design based on IDT xGen Exome Research Panel v1.0 (Fulgent Genetics), and xGen Hybrdization and Wash Kit (Integrated DNA Technologies). Enriched samples were sequenced with 2×150bp on an Illumina NovaSeq6000 (Illumina Inc).
2.4. Bioinformatics analysis
After sequencing, the reads were obtained and aligned to assembly hg19 of the human genome using NextGENe V2.3.4. Base quality recalibration and local realignment were performed by the Genome Analysis Tool Kit. Variants were called using NextGENe V2.3.4. Variant frequency data were obtained from public and internal control databases including the Exome Aggregation Consortium (ExAC) database, dbSNP database, ClinVar database, the Human Gene Mutation Database (HGMD) and 1000 Genomes Project. Variants with a frequency higher than 0.5% were filtered out. Annotate Variation (ANNOVAR) was used to annotate protein-altering changes and dbNSFP (contains SIFT, PolyPhen-2, LRT, Mutation Taster) was used to predict the pathogenicity about the deleteriousness of variants.
2.5. Sanger sequencing and segregation analysis
Sanger sequencing was performed to validate the variants identified by WES. Polymerase chain reactions (PCR) primer pairs were designed on Primer 3 software (http://bioinfo.ut.ee/primer3-0.4.0/) for each exon of interest. After PCR amplification, the amplicons were sequenced on an Applied Biosystems 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequences were assembled and analyzed using Lasergene SeqMan software (DNASTAR, Madison, WI, USA). All available family members were Sanger sequenced in order to perform segregation tests.
2.6. RNA analysis
Total RNAs were extracted from the periphery blood samples of the patients and unaffected control subjects using the QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Single-stranded cDNA was synthesized from RNA using GoScript Reverse Transcription System (Promega, Madison, USA) according to manufacturer’s protocol. RT-PCR primers were designed on Primer 3 software to amplify the segments encompassing splicing variants. After PCR amplification, the amplicons were sequenced on an Applied Biosystems 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequences were assembled and analyzed using Lasergene SeqMan software (DNASTAR, Madison, WI, USA).
2.7. Statistical analysis
All patients with biallelic PNPLA6 variants in the reported literatures and this study were enrolled to take genotype-phenotype analysis. Chi-square test and Fisher’s exact test were applied to compare the differences between ocular involved patient group and the without ocular phenotype group using the SPSS version 19.0. P value < 0.05 was considered statistically significant.
3. Results
3.1. Clinical findings
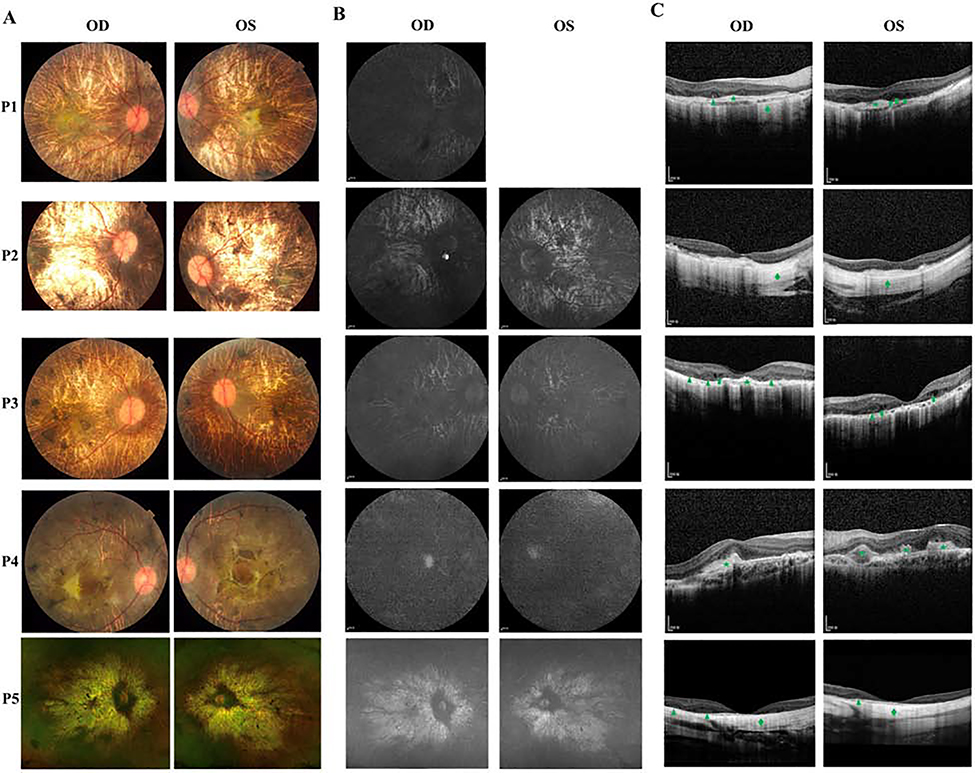
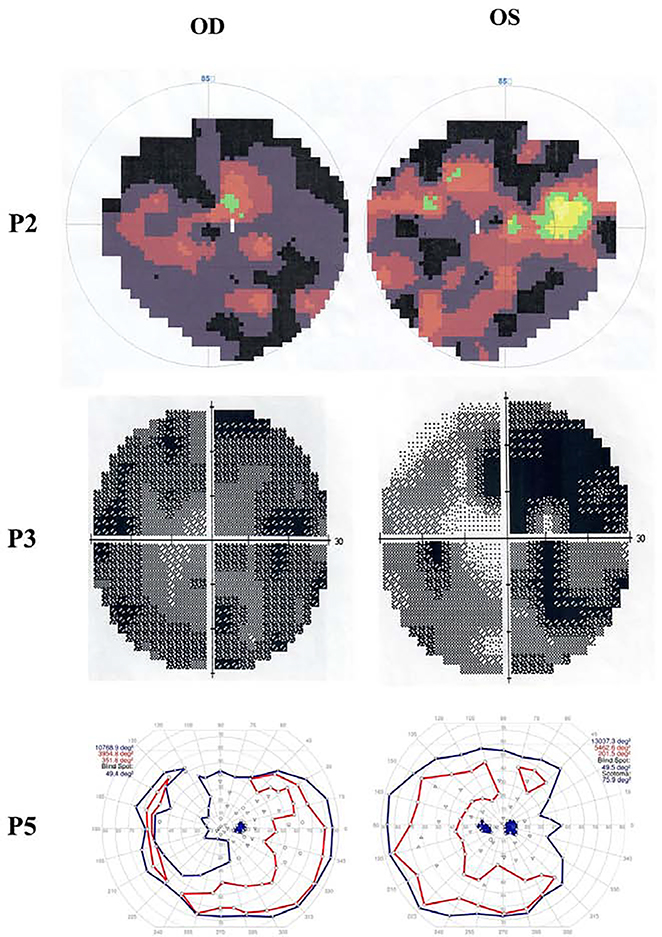
A total 3 males and 1 female from 4 unrelated Chinese families and 1 female with mixed Caucasian/Chinese background are recruited for this study (Figure 1A). The detailed clinical findings of 5 patients were summarized in Table 1. The mean age of patients was 20.8 years (range, 4–28). The mean age at first visit was 14.2 years (range, 7–25). P1, P2, P3 and P5 were followed for 5, 4, 3 and 21 years, respectively. All patients had night blindness and vision impairment since childhood. BCVA ranged from 20/200 to 20/2000. Color fundus photography revealed severe chorioretinal dystrophy with scattered pigment clumps or areas of chorioretinal dystrophy with visible sclera (Figure 2A). Fundus autofluorescence (FAF) photographs displayed extensive hypofluorescence (Figure 2B). OCT indicated disorganization and thinning of outer retina, outer retinal tubulations, loss of external limiting membrane, ellipsoid and interdigitation zone, thinning of the retinal pigment epithelium (RPE) and choriocapillaris (Figure 2C). Full-field ERGs were recorded in four patients (P1, P2, P4, P5) and three patients (P1, P2, P3) showed extinguished scotopic and photopic responses and P5 had remarkably reduced cone and rod responses at age 9. Three patients (P2, P3, P5) were able to take visual field test and two (P2, P3) presented with severe visual field defects (Figure 3). Visual fields of P2 indicated tunnel vision (OD) and residual 5° central and upper nasal quadrants (OS). Visual fields of P3 remained about 2° (OD) and 5° (OS) using a size Ⅲ, white stimulus. Visual fields in P5 between the ages of 20 and 25 demonstrated intact responses to a V4e target peripherally but variably responses centrally, often demonstrating a scotoma to this target OU. Obvious systemic symptoms were observed in four patients (P1, P2, P3, P5) during the follow-up as described below.
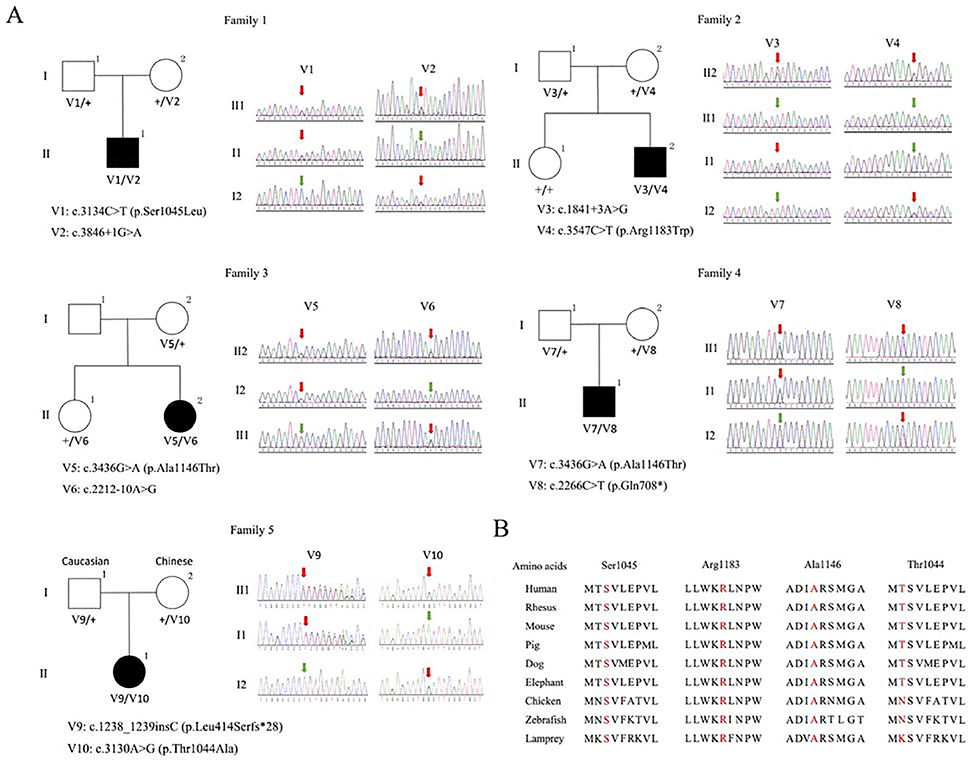
Figure 1.
A: Pedigrees and Sanger sequencing chromatograms of PNPLA6-related disorders patients in this study. A total of five unrelated patients with PNPLA6 biallelic variants were identified. Variant annotations were based on GenBank: NM_001166111. Squares indicate men; circles indicate women; black indicates patients; red arrows indicate the variants; green arrows indicate the normal nucleotide. B: Conservative analysis of four amino acids affected by missense variants. The Thr1044, Ser1045, Ala1146 and Arg1183 are highlighted and evolutionarily conserved.
Table 1.
Summary of the clinical manifestations of five patients
| Patient | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|
| Gender | M | M | F | M | F |
| Ethnicity | Han Chinese | Han Chinese | Han Chinese | Han Chinese | Caucasian/Chinese |
| Age (years) | 12 | 28 | 28 | 11 | 25 |
| Age at initial visit (years) | 7 | 24 | 25 | 11 | 4 |
| Phenotype | OMS | BNS | BNS | BNS | BNS |
| BCVA(OD,OS) | 20/2000,20/630 | 20/200,20/200 | 20/500,20/500 | 20/200,20/250 | 20/600,20/1000 |
| Night blindness | + | + | + | + | + |
| Chorioretinopathy | + | + | + | + | + |
| ERG | No recordable responses | No recordable responses | NA | No recordable responses | Remarkably reduced |
| Ataxia | − | + | − | − | − |
| MRI of brain | small pituitary gland | Atrophy cerebellum | − | − | NA |
| Hypogonadotropic hypogonadism | + | + | + | + | + |
| Testosterone | Low | Low | Normal | Slightly low | NA |
| GH deficiency | + | NA | NA | − | NA |
| Short stature | + | − | − | − | + |
| Delayed bone age | + | NA | NA | NA | NA |
| Trichomegaly | + | − | − | − | − |
| Sparse hair | + | − | − | − | − |
M, male; F, female; OMS, Oliver-McFarlane syndrome; BNS, Boucher–Neuhäuser syndrome; BCVA, best corrected visual acuity; MRI, magnetic resonance imaging; ERG, electroretinography; GH, growth hormone; NA, not available;
Figure 2.
A: Color fundus photographs of five patients. P1, P2, P3 and P5 all presented extensive chorioretinal dystrophy and visualization of sclera with pigment clumps. P4 presented chorioretinal dystrophy, scattered pigmentation and fibrosis patches. B: The FAF photographs of five patients. All displayed generalized remarkably decreased autofluorescence. C: OCT images of five patients. All revealed disorganization and thinning of outer retina, loss of external limiting membrane, ellipsoid and interdigitation zone, thinning of the RPE and choriocapillaris. P1 and P3 had outer retinal hyporeflective cysts (arrow). P1, P3, and P5 displayed outer retinal tubulations (arrowhead). P1, P3 and P4 indicated hyperreflective retinal fibrosis (star). P1, P2 and P5 showed hyperreflective scleral signals (diamond).
Figure 3.
Visual field images of P2, P3 and P5. P2 displayed tunnel vision (OD) and residual 5°central and upper nasal quadrants (OS). P3 remained about 2°(OD) and 5°(OS) tunnel visual field. P5 had intact responses to V4e target peripherally but decreased responses to the III4e and smaller targets centrally (OU).
P1 was 7 years old when he first presented complaining of decreased visual acuity and nyctalopia. He was diagnosed possibly having choroideremia or X-linked retinitis pigmentosa, but no causative variants were identified in CHM, RPGR or RP2. At 11 years old, he was diagnosed with short stature (131cm). Hormonal testing revealed a deficiency of growth hormone (GH), insulin like growth factor 1 (IGF1) and testosterone; parathyroid hormone (PTH) was high and thyroid stimulating hormone (TSH) was normal. The radiological bone age was also delayed, and MRI of the brain showed small pituitary gland. In addition, he was observed to have long eyelashes, eyebrows and sparse hair. During the 5-year follow-up, the visual acuity decreased from 20/200 (OD) and 20/100 (OS) to 20/2000 (OD) and 20/630 (OS).
Patient P2 presented at 24 years old with complaint of blurred vison and night blindness since childhood. The fundus showed extensive chorioretinopathy and the diagnosis of choroideremia was made. However, panel capture sequencing and MLPA did not find a disease-causing variant in CHM. He developed a tremor of the hands and head at 26 years old. Further investigation revealed he had delayed secondary sexual development with testosterone deficiency at 20 years old and testosterone replacement therapy was initiated. MRI of the brain indicated atrophy of the cerebellar hemispheres. During the 4-year follow-up, the visual acuity 20/200 (OU) was stable.
Patient P3 was referred at age 25 with a diagnosis of retinitis pigmentosa and reported vision loss and night blindness for more than 10 years. Her past medical history included primary amenorrhea. Hormonal testing revealed luteinizing hormone (LH), follicle-stimulating hormone (FSH), estradiol (E2) and prolactin (PRL) were significantly lower than normal. Ultrasound of the abdomen indicated hypoplasia of uterus. During the 3-year follow-up, there was no significant change in vision (20/500, OU).
P4 reported a three-year history of nyctalopia and decreased visual acuity and with a rapid loss of visual acuity over the previous 3 months. At presentation, his visual acuity was 20/200 (OD) and 20/250 (OS). After PNPLA6 compound heterozygous variants were confirmed, hormonal testing was obtained and showed testosterone was 1.68ng/ml, slightly lower than normal (1.75–7.81ng/ml), while other hormones were normal.
P5 presented at the age of 4 with a history of nyctalopia. Full field ERGs revealed a rod-cone dystrophy and a diagnosis of early onset severe retinal dystrophy (EOSRD) was made. At age 12, she was diagnosed with short stature (below the 5th percentile) and at age 14 with hypogonadotrophic hypogonadism and started at on estrogen replacement therapy. As of age 25 she had not manifested any neurological symptoms. Genetic testing revealed compound heterozygous variants in PNPLA6.
3.2. Genetic findings
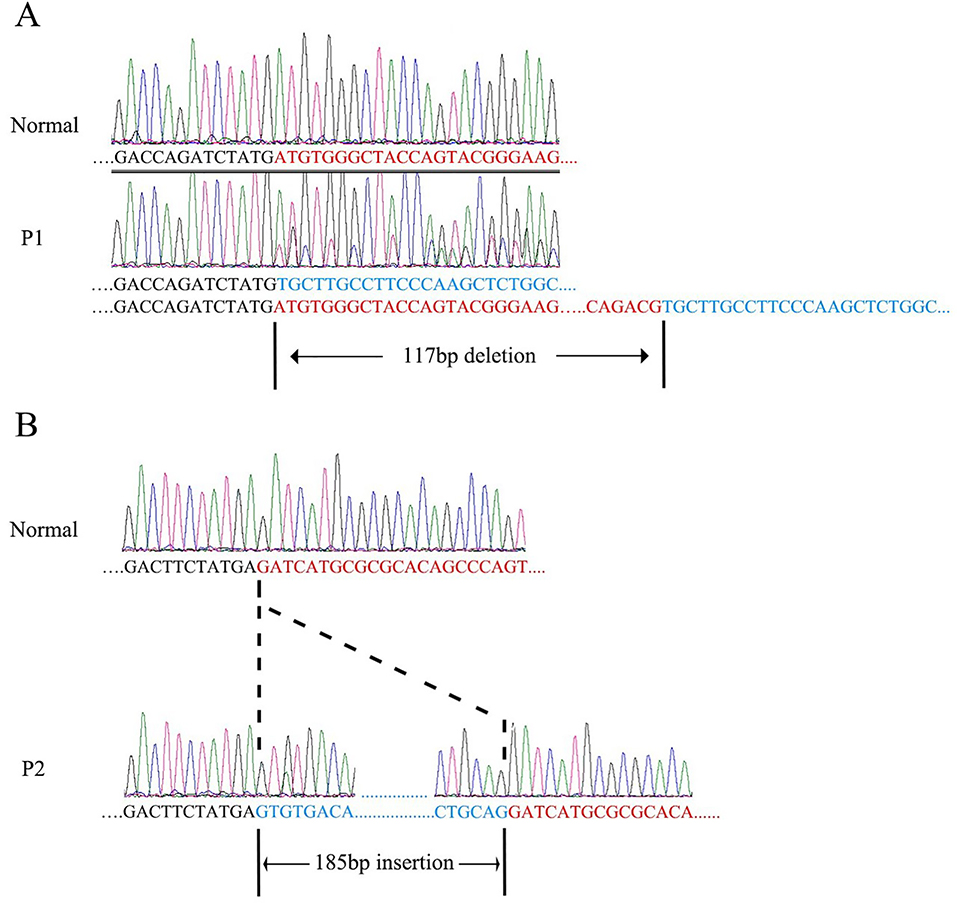
We performed WES and obtained high quality data (mean coverage: 120×) for exonic regions. After filtering and prioritization, WES data revealed that biallelic pathogenic variants in PNPLA6 were identified in all five patients (annotation according to GenBank: NM_001166111). There was one insertion, one nonsense, three splicing and four missense variants found in five patients. Compound heterozygous c.3134C>T (p.Ser1045Leu) and c.3846+1G>A, c.3547C>T (p.Arg1183Trp) and c.1841+3A>G, c.3436G>A (p.Ala1146Thr) and c.2212–10A>G, c.3436G>A (p.Ala1146Thr) and c.2266C>T (p.Gln756*), c.1238_1239insC (p.Leu414Serfs*28) and c.3130A>G (p.Thr1044Ala) were identified in P1, P2, P3, P4 and P5, respectively. Sanger sequencing was performed to validate the variants and confirm the co-segregation of variants with disease phenotype in the pedigrees (Figure 1A). Three variants (c.3134C>T, p.Ser1045Leu; c.3547C>T, p.Arg1183Trp; c.1238_1239insC, p.Leu414Serfs*28) were reported previously(Synofzik et al., 2014; Stone et al., 2017; Kmoch et al., 2015), and six variants (c.2266C>T, p.Gln708*; c.3436G>A, p.Ala1146Thr; c.3130A>G, p.Thr1044Ala; c.1841+3A>G; c.3846+1G>A and c.2212–10A>G) were novel. All of the missense variants are well conserved across different species (Figure 1B). To further confirm that the identified PNPLA6 splicing variants were bona fide splicing-disrupting, RT-PCR and Sanger sequencing were performed. For the splicing variant c.3846+1G>A, a 117bp deletion of PNPLA6 mRNA (r.3731_3847del) was noted (Figure 4A). For the splicing variant c.1841+3A>G, a 185bp insertion of PNPLA6 mRNA (r.1841_1842ins1841+1_1841+185) was detected (Figure 4B). The mRNA of P3 was unavailable. SpliceSiteFinder-like, MaxEntScan, Human Splicing Finder all predicted the splicing variant c.2212–10A>G would influence splice. The results of genetic analysis were summarized in Table 2.
Figure 4.
RT-PCR and chromatograms of P1 (A) and P2 (B). A: A 117bp region was deleted in mRNA compared with normal control. B: A 185bp region was inserted in mRNA compared with normal control.
Table 2.
Summary of the variants analysis of five patients
| Patient | Diagnosis | DNA change | Protein/mRNA change | Variant type | SIFT | Polyphen2 | LRT | Mutation Taster | Novel |
|---|---|---|---|---|---|---|---|---|---|
| P1 | OMS | c. 3134C>T | p.Ser1045Leu | Missense | - | - | - | - | N |
| c.3846+1G>A | r.3711_3847del | Splicing | - | - | - | - | Y | ||
| P2 | BNS | c. 3547C>T | p.Arg1183Trp | Missense | - | - | - | - | N |
| c.1841+3A>G | r.1841_1842ins1841+1_1 841+185 |
Splicing | - | - | - | - | Y | ||
| P3 | BNS | c.3436G>A | p.Ala1146Thr | Missense | T | PD | D | DC | Y |
| c.2212–10A>G | NA | Splicing | - | - | - | - | Y | ||
| P4 | BNS | c.3436G>A | p.Ala1146Thr | Missense | T | PD | D | DC | Y |
| c.2266C>T | p.Gln756* | Nonsense | - | - | - | DC | Y | ||
| P5 | BNS | c.1238_1239insC | p.Leu414Serfs*28 | Insertion | - | - | - | - | N |
| c.3130A>G | p.Thr1044Ala | Missense | T | B | D | DC | Y |
OMS, Oliver-McFarlane syndrome; BNS, Boucher–Neuhäuser syndrome; T, tolerated; B, benign; PD, probably damaging; D, deleterious; DC, disease causing; Y, yes; N, no; NA, not available. The splicing variant c.2212–10A>G would influence splice predicted by SpliceSiteFinder-like, MaxEntScan, Human Splicing Finder. All the variants are described according to PNPLA6 transcript variant (NM_001166111)
3.3. Genotype–phenotype analysis
In total, 70 affected subjects (65 reported in the literature and 5 in this study) from 49 pedigrees harboring biallelic pathogenic PNPLA6 variants were ascertained from previous literature and this study. Among 49 families, 34 (34/49, 69.4%) had chorioretinal dystrophy and 15 (15/49, 30.6%) had no reported ocular phenotype. The genotype/phenotype correlation of these PNPLA6 cases are summarized in Table 3. A total 98 alleles and 71 different variants are also analyzed. The variants are diverse and widely distributed in the gene. Only 2 variants, c.3084_3085insGCCA and p.Gly1129Arg are considered as frequent variants, accounting for 9.18% (9/98) and 5.10% (5/98), respectively. The novel variant p.Ala1146Thr identified in this study, accounts for 20% (2/10) in this study, and was the most frequent allele.
Table 3.
List of all available PNPLA6 variants and associated phenotypes
| Reference | Total case | Eye involved | Phenotype | Variant 1 | Effect 1 | Variant type | Domain 1 | Variant 2 | Effect 2 | Variant type | Domain 2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Zheng et al, 2018 | 1 | Y | BNS | c.3386G>T | p.G1129V | Missense | Pat | c.3534G>C | p.W1178C | Missense | Pat |
| Tarnutzer et al, 2015 | 1 | Y | BNS | c.288T>G | p.Y96* | Nonsense | - | c.865C>G | p.R289G | Missense | cNMP |
| Tarnutzer et al, 2015 | 1 | Y | BNS | c.343–2A>T | - | Splicing | - | c.4075C>T | p.R1359W | Missense | - |
| Synofzik et al, 2014 | 2 | Y | BNS | c.2212–1G>C | p.V738Qfs*98 | Splicing | cNMP | c.3328G>A | p.V1110M | Missense | Pat |
| Synofzik et al, 2014 | 1 | N | Sporadic ataxia | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.3299T>G | p.V1100G | Missense | Pat |
| Synofzik et al, 2014 | 1 | N | HSP | c.787G>A | p.V263I | Missense | cNMP | c.2519G>A | p.G840E | Missense | - |
| Synofzik et al, 2014 | 1 | Y | BNS | c.3134C>T | p.S1045L | Missense | Pat | c.3365C>T | p.P1122L | Missense | Pat |
| Synofzik et al, 2014 | 2 | Y | BNS | c.1732G>T | p.G578W | Missense | cNMP | c.3197T>C | p.F1066S | Missense | Pat |
| Synofzik et al, 2014 | 1 | N | GHS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.4084C>G | p.R1362G | Missense | - |
| Synofzik et al, 2014 | 4 | Y | BNS | c.3173C>T | p.T1058I | Missense | Pat | c.3173C>T | p.T1058I | Missense | Pat |
| Kmoch et al, 2015 | 1 | Y | OMS | c.1571T>C | p.L524P | Missense | cNMP | c.3373G>A | p.D1125N | Missense | Pat |
| Kmoch et al, 2015 | 1 | Y | OMS | c.2116C>T | p.Q706* | Nonsense | cNMP | c.3385G>C | p.G1129R | Missense | Pat |
| Kmoch et al, 2015 | 3 | Y | OMS | c.343–2A>T | - | Splicing | - | c.3373G>A | p.D1125N | Missense | Pat |
| Kmoch et al, 2015 | 1 | Y | LCA | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.1076C>T | p.T359I | Missense | - |
| Kmoch et al, 2015 | 1 | Y | OMS | c.3322C>T | p.R1108W | Missense | Pat | c.3385G>C | p.G1129R | Missense | Pat |
| Kmoch et al, 2015 | 1 | Y | OMS | c.1238_1239insC | p.L414Sfs*28 | Insertion | - | c.3385G>C | p.G1129R | Missense | Pat |
| Fogel et al, 2014 | 1 | N | Cerebellar ataxia | c.1484C>T | p.P495L | Missense | - | c.3742C>G | p.Q1248E | Missense | Pat |
| Coutelier et al, 2018 | 1 | N | Spastic ataxia | c.2633G>A | p.G878D | Missense | - | c.2633G>A | p.G878D | Missense | - |
| Coutelier et al, 2018 | 1 | N | Cerebellar ataxia | c.1483C>A | p.P495T | Missense | - | c.1857G>T | p.Q619H | Missense | cNMP |
|
Rainier et al, 2008
Rainier et al, 2011 |
2 | N | MND | c.3178A>G | p.M1060V | Missense | Pat | c.3178A>G | p.M1060V | Missense | Pat |
|
Rainier et al, 2008
Rainier et al, 2011 |
2 | N | MND | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.2813G>A | p.R938H | Missense | - |
| Yoon et al, 2013 | 1 | N | Spastic paraplegia | c.1816_1818delinsTA | p.Arg606* | Indels | cNMP | del exons 16–17 | - | Deletion | cNMP |
| Hufnagel et al, 2015 | 2 | Y | OMS/LMS | c.3296G>A | p.R1099Q | Missense | Pat | c.3526G>A | p.G1176S | Missense | Pat |
| Hufnagel et al, 2015 | 1 | Y | OMS/LMS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.3385G>A | p.G1129R | Missense | Pat |
| Hufnagel et al, 2015 | 1 | Y | OMS/LMS | c.1973+2T>G | - | Splicing | cNMP | c.3644T>C | p.V1215A | Missense | Pat |
| Hufnagel et al, 2015 | 1 | Y | OMS/LMS | Dup exons 14–20 | - | Insertion | cNMP | c.3644T>C | p.V1215A | Missense | Pat |
| Hufnagel et al, 2015 | 1 | Y | OMS/LMS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.3385G>A | p.G1129R | Missense | Pat |
| Hufnagel et al, 2015 | 4 | Y | OMS/LMS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.2176G>C | p.G726R | Missense | cNMP |
| D’Amore et al, 2018 | 1 | N | Spastic paraplegia | c.3585C>G | p.D1195E | Missense | Pat | c.2389G>A | p.V797M | Missense | - |
| Teive et al, 2018 | 1 | N | GHS | c.4081C>T | p.R1361* | Missense | - | c.3373G>A | p.D1125N | Missense | Pat |
| Teive et al, 2018 | 1 | Y | BNS | c.3531G>A | p.W1177* | Missense | Pat | c.2441T>C | p.L814P | Missense | - |
| Koh et al, 2015 | 1 | Y | BNS | c.3523_3524insTGTCCG | p.1175_1176insVS | Insertion | Pat | c.2923A>G | p.T975A | Missense | Pat |
| Koh et al, 2015 | 1 | Y | BNS | c.3534G>C | p.W1178C | Missense | Pat | c.3534G>C | p.W1178C | Missense | Pat |
| Deik et al, 2014 | 1 | Y | BNS | c.3134C>T | p.S1045L | Missense | Pat | c.3519C>G | p.S1173R | Missense | Pat |
| Stone et al, 2017 | 1 | Y | Retinal dystrophy | c.3334G>A | p.A1112T | Missense | Pat | c.3547C>T | p.R1183W | Missense | Pat |
| Topaloglu et al, 2014 | 2 | N | GHS | c. 3524C>G | p.S1175C | Missense | Pat | c. 3524C>G | p.S1175C | Missense | Pat |
| Topaloglu et al, 2014 | 2 | Y | BNS | c.1270_1271insG | p.D424Gfs*18 | Insertion | - | c.3439C>T | p.R1147C | Missense | Pat |
| Topaloglu et al, 2014 | 2 | N | GHS | c.2494_2495ins13 | p.G832Vfs*27 | Insertion | - | c.4075C>T | p.R1359W | Missense | - |
| O’Neil et al, 2019 | 1 | Y | BNS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.3548G>A | p.R1183Q | Missense | Pat |
| Patsi et al, 2018 | 1 | Y | OMS | c.3620C>A | p.S1207Y | Missense | Pat | c.3620C>A | p.S1207Y | Missense | Pat |
| #Wiethoff et al, 2017 | 2 | N | Cerebellar ataxia | c.3929A>T/ c.3847G>A | p.D1310V/ p.V1283M |
Missense/ Missense |
-/- | c.3929A>T/ c.3847G>A |
p.D1310V/ p.V1283M |
Missense/ Missense |
-/- |
| Langdahl et al, 2017 | 2 | Y | BNS | c.3084_3085insGCCA | p.R1031Qfs*38 | Insertion | Pat | c.4076G>A | p.R1359Q | Missense | - |
| Salgado et al, 2019 | 4 | N | GHS | c.4081C>T | p.R1361* | Missense | - | c.2404G>C | p.E802Q | Missense | - |
| DeNaro et al, 2018 | 1 | Y | BNS | c.1431T>A | p.C477* | Nonsense | - | c.3034G>A | p.G1012S | Missense | Pat |
| This study | 1 | Y | OMS | c.3846+1G>A | r.3711_3847del | Splicing | Pat | c. 3134C>T | p.S1045L | Missense | Pat |
| This study | 1 | Y | BNS | c.1841+3A>G | r.1841_1842ins1841 +1_1841+185 |
Splicing | cNMP | c. 3547C>T | p.R1183W | Missense | Pat |
| This study | 1 | Y | BNS | c.2212–10A>G | - | Splicing | cNMP | c.3436G>A | p.A1146T | Missense | Pat |
| This study | 1 | Y | BNS | c.2266C>T | p.Q756* | Nonsense | - | c.3436G>A | p.A1146T | Missense | Pat |
| This study | 1 | Y | BNS | c.1238_1239insC | p.L414Sfs*28 | Insertion | - | c.3130A>G | p.T1044A | Missense | Pat |
| Total | 70 |
cNMP: cyclic nucleotide monophosphate (cNMP) binding domain (amino acid interval, 195–316/512–622/635–743); Pat: patatin-like phospholipase domain (amino acid interval, 964–1269); BNS: Boucher–Neuhäuser syndrome; HSP: hereditary spastic paraplegia; GHS: Gordon Holmes syndrome; OMS: Oliver-McFarlane syndrome; LMS: Laurence–Moon syndrome; LCA: Leber congenital amaurosis; MND: motor neuron disease; Y: Yes; N: No
two homozygous missense variants in this consanguineous family; All the variants are described according to PNPLA6 transcript variant (NM_001166111)
Among 71 variants, there are 53 missense variants (53/71, 74.65%), which is the most common variant type, followed by splicing variants (6/71, 8.45%), small insertion and indel variants (6/71, 8.45%), nonsense variants (4/71, 5.63%), and gross insertion and deletion variants (2/71, 2.82%). We classified splicing, nonsense, small and gross insertion and deletion variants as severe allele and missense variant as mild allele. Based on this, we categorized the patients into two groups: those with one or two severe alleles and those with only mild alleles. We found that patients with chorioretinal dystrophy carried more severe deleterious variants (22/34, 64.7%) than patients without chorioretinal dystrophy (5/15, 33.3%), P<0.05 by Chi-square test. Only one patient carrying two severe deleterious alleles presented with spastic paraplegia.
Among 53 missense variants, 32 (32/53, 60.4%) of them are located in patatin-like phospholipase (Pat) domain (amino acid interval, 964–1269) and 6 (6/53, 11.3%) in cyclic nucleotide monophosphate (cNMP) binding domain (amino acid interval, 195–316/512–622/635–743). Among 18 severe variants, 3 (3/18, 16.7%) are located in Pat domain and 8 (8/18, 44.4%) in cNMP binding domain. More subjects with chorioretinal dystrophy (9/34, 26.5%) harbor biallelic variants in both domains (one variant in Pat domain and the other one in cNMP binding domain) than that of subjects without chorioretinal dystrophy (0/15, 0%), but the result was not statistically significant (P=0.071 by Chi-square test). We also discovered that patients with chorioretinal dystrophy carried significantly more variants (32/34, 94.12%) in Pat domains (≥1 variant in Pat domain) than those without retinal phenotypes (8/15, 53.3%) (P<0.01 by Chi-square test). Another interesting finding is that 3 splicing variants were identified in our 4 Chinese patients, but only 4 splicing alleles were found previously in 44 families.
4. Discussion
PNPLA6 belongs to a family of nine patatin-like phospholipase domain-containing proteins. It is mainly expressed in the brain and involved with neural development. Deficits are associated with neurodegeneration. It is also expressed in several non-neuronal tissues such as testes and kidney (Sogorb et al., 2016; Winrow et al., 2003; Hufnagel et al., 2015). PNPLA6 is expressed in the retina, crystal lens, pituitary, cerebellum and ventricular zones of the brain (Hufnagel et al., 2015). Knockdown of pnpla6 expression resulted in developmental abnormalities and motor neuron defects in zebrafish (Hufnagel et al., 2015; Song et al., 2013). Swiss cheese protein (the orthologue of vertebrate PNPLA6) was essential for membrane lipid homeostasis and cell survival in both neurons and glia of the adult Drosophila brain. It was proposed that NTE might play an analogous role in vertebrates(Mühlig-Versen et al., 2005). Furthermore, it was found that PNPLA6 was expressed in horizontal, amacrine and photoreceptors cells, which was crucial for photoreceptor maintenance in adult Drosophila. PNPLA6 is localized mostly in the plasma membrane of the inner segments of mouse photoreceptors, suggesting its role in the formation of distinct plasma membrane domains in the photoreceptors or maintenance of photoreceptor integrity(Kmoch et al., 2015).
All of our patients manifested vision loss and night blindness at very young age without any obvious systemic abnormalities. They were diagnosed with non-syndromic retinal degeneration (EOSRD, choroideremia or retinitis pigmentosa) until systemic involvement became apparent or genetic testing was performed. We followed four patients (P1, P2, P3, P5) for a number of years and found that they had varying degrees of progressive vision loss. In our series, all patients presented with a distinct chorioretinal atrophy. However, the retinal appearance of PNPLA6-related disorders can be diverse, with the full spectrum ranging from a mild retinal pigment epitheliopathy to severe chorioretinal atrophy(Zheng et al., 2018; Salvador et al., 1995; DeNaro et al., 2018; Teive et al., 2018). The degeneration can be confined to the macula, mid periphery(Tarnutzer et al., 2015), or involve the entire retina and mimic advanced choroideremia(Yu et al., 2008; Kmoch et al., 2015; O’Neil et al., 2019; Synofzik et al., 2014; Deik et al., 2014). Since systemic features many not present later in life, it is important to consider the possibility of PNPLA6-related disorders in patients with choroideremia-like changes, especially when genetic testing is inconclusive.
For patients found to have PNPLA6-associated syndrome, a full systemic work up is critical. This should include a referral to an endocrinologist and neurologist. Four patients (P2, P3, P4, P5) were diagnosed as Boucher–Neuhäuser syndrome (BNS) and P1 was diagnosed as Oliver-McFarlane syndrome (OMS) based on their clinical assessments and genetic data. In our series, three adult patients (P2 ,P3, P5) exhibited hypogonadotropic hypogonadism. However, hormone replacement therapy can be effective if hormone abnormalities are detected in adolescence(Boehm et al., 2015).
The PNPLA6-related disorders are a complex group of diseases including several syndromes. Possible genotype-phenotype correlation had been proposed in previously, but not confirmed (Synofzik et al., 2014; Kmoch et al., 2015). Synofzik et al observed that variants in the N-terminal side of the Pat domain might associate with spasticity (spastic paraplegia and spastic ataxia), while variants towards the C-terminal end of the Pat domain might associate with cerebellar ataxia and/or hypogonadism (Boucher-Neuhäuser syndrome, Gordon Holmes syndrome, spastic ataxia) based on 7 families(Synofzik et al., 2014). Meanwhile, Kmoch et al noticed that biallelic variants in Pat and cNMP binding domains could cause a more severe impairment of PNPLA6 function and lead to photoreceptor death based on 7 families(Kmoch et al., 2015).
However, the above putative phenotype-genotype correlation was obtained from limited research cases. After dissecting the genotypes and phenotypes of all PNPLA6 patients, we bring out some perspectives regarding connections between genotype and phenotype. First, our data suggests that variant position may influence phenotype. Patients with chorioretinal dystrophy tended to have PNPLA6 variants located in both domains (one in Pat domain and the other one in cNMP binding domain) as Kmoch et al detected(Kmoch et al., 2015). However, this finding was not statistically significant (P=0.071). Nevertheless, we demonstrate that Pat domain plays the key role instead of Pat domain plus cNMP binding domain. One allele or biallelic variants in Pat domain is highly correlated with the chorioretinal dystrophy (P<0.01). Second, variant type may also relate to phenotype. More PNPLA6 patients with severe deleterious variants presented with chorioretinal dystrophy (P<0.05). However, one patient with two severe deleterious variants presented only with spastic paraplegia without retinopathy (Yoon et al., 2013), which is contrary to this conclusion from a dosage standpoint. In addition, we presume that ethnical specific alleles may exist. The c.3084_3085insGCCA (9/87, 10.34%) and p.Gly1129Arg (5/87, 5.75%) are recurrent alleles in Caucasians. While in Chinese, variant p.Ala1146Thr (2/11, 18.18%) is the most common one and may be a mutational hot spot. Furthermore, it seemed that splicing variants were much more frequent in Chinese (3/11, 27.27%) than that of Caucasians (4/87, 4.60%) (P<0.05 by Fisher’s exact test). As the Chinese sample size is very limited, more data are needed to draw a conclusion. Notwithstanding the genotype-phenotype correlations, intra-familial phenotypic variation was reported. For example, two siblings in their 40s, with the same biallelic variants, one had BNS with chorioretinal dystrophy and the other one didn’t show any evidence of chorioretinal dystrophy(Synofzik et al., 2014).
Most of the identified PNPLA6 variants (35/71, 49.3%) were located within the Pat domain, critical for the esterase activity of NTE. This domain has been shown to de-esterify phosphatidylcholine, a major component of biological membranes, into its constituent fatty acids and glycerophosphocholine (Synofzik et al., 2014). Several studies indicated the loss of NTE enzymatic activity of the Pat domain was closely correlated to the PNPLA6-related disorders(Synofzik et al., 2014; Hufnagel et al., 2015; Hein et al., 2010a; Hein et al., 2010b). Structural modeling and analysis revealed that the location of variants in the Pat domain was associated with NTE activity(Synofzik et al., 2014; Hufnagel et al., 2015). In addition, it was suggested that the onset and severity of disease was related to the hydrolase activity of the Pat domain, contributing to the phenotypic heterogeneity(Hufnagel et al., 2015). These data support our finding that chorioretinal dystrophy is highly correlated with the variants in Pat domain. In addition, Chang et al. revealed that PNPLA6 exhibited dynamic interactions with the endoplasmic reticulum and lipid droplets that depended on the interplay of two functional regions, the amino-terminal region and the carboxyl-terminal catalytic region. Variants that disrupt this interplay may contribute to PNPLA6-related disorders by affecting protein positioning(Chang et al., 2019).
In summary, PNPLA6-related disorder is a group of diseases with miscellaneous and overlapping clinical features. Choroideremia-like retinal changes are the characteristic ocular phenotype, which can appear as the only finding at an early age. Chorioretinal dystrophy correlates with variant type and variants in Pat domain. Whole exome sequencing is a useful tool to elicit the genetic diagnosis. This paper is the most detailed report and review regarding PNPLA6-related chorioretinal dystrophy so far. Our findings expand the clinical and genetic spectrum related to the rare inherited diseases.
The paper described phenotype and genetic defects of 5 PNPLA6-related syndromic retinal dystrophy patients from 5 unrelated families.
Choroideremia-like retinal changes are the characteristic ocular phenotype of PNPLA6-related syndromic retinal dystrophy.
Genotype-phenotype analysis found chorioretinal dystrophy correlates with variant type and variants in Pat domain.
Acknowledgments
The authors thank the patients who participated in the study. This work was supported by the National Natural Science Foundation of China (81873687), Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (CIFMS 2016-12M-1-002), unrestricted grant from Research to Prevent Blindness (Casey Eye Institute), P30 Core Grant NIH P30EY010572 (Casey Eye Institute).
Footnotes
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- Boehm U, Bouloux PM, Dattani MT, de Roux N, Dode C, Dunkel L, Dwyer AA, Giacobini P, Hardelin JP, Juul A, Maghnie M, Pitteloud N, Prevot V, Raivio T, Tena-Sempere M, Quinton R Young J, 2015. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism--pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 11: 547–564. [DOI] [PubMed] [Google Scholar]
- Chang P, He L, Wang Y, Heier C, Wu Y Huang F, 2019. Characterization of the Interaction of Neuropathy Target Esterase with the Endoplasmic Reticulum and Lipid Droplets. Biomolecules 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelier M, Hammer MB, Stevanin G, Monin ML, Davoine CS, Mochel F, Labauge P, Ewenczyk C, Ding J, Gibbs JR, Hannequin D, Melki J, Toutain A, Laugel V, Forlani S, Charles P, Broussolle E, Thobois S, Afenjar A, Anheim M, Calvas P, Castelnovo G, de Broucker T, Vidailhet M, Moulignier A, Ghnassia RT, Tallaksen C, Mignot C, Goizet C, Le Ber I, Ollagnon-Roman E, Pouget J, Brice A, Singleton A Durr A, 2018. Efficacy of Exome-Targeted Capture Sequencing to Detect Mutations in Known Cerebellar Ataxia Genes. JAMA Neurol 75: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amore A, Tessa A, Casali C, Dotti MT, Filla A, Silvestri G, Antenora A, Astrea G, Barghigiani M, Battini R, Battisti C, Bruno I, Cereda C, Dato C, Di Iorio G, Donadio V, Felicori M, Fini N, Fiorillo C, Gallone S, Gemignani F, Gigli GL, Graziano C, Guerrini R, Gurrieri F, Kariminejad A, Lieto M, Marques LourenCo C, Malandrini A, Mandich P, Marcotulli C, Mari F, Massacesi L, Melone MAB, Mignarri A, Milone R, Musumeci O, Pegoraro E, Perna A, Petrucci A, Pini A, Pochiero F, Pons MR, Ricca I, Rossi S, Seri M, Stanzial F, Tinelli F, Toscano A, Valente M, Federico A, Rubegni A Santorelli FM, 2018. Next Generation Molecular Diagnosis of Hereditary Spastic Paraplegias: An Italian Cross-Sectional Study. Front Neurol 9: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deik A, Johannes B, Rucker JC, Sánchez E, Brodie SE, Deegan E, Landy K, Kajiwara Y, Scelsa S, Saunders-Pullman R Paisán-Ruiz C, 2014. Compound heterozygous PNPLA6 mutations cause Boucher-Neuhäuser syndrome with late-onset ataxia. J Neurol 261: 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNaro BB, Dhrami-Gavazi E, Rubaltelli DM, Freund KB, Lee W, Yannuzzi LA, Tsang SH Kang JJ, 2018. CHORIORETINAL CHANGES IN A GENETICALLY CONFIRMED CASE OF BOUCHER-NEUHÄUSER SYNDROME. Retin Cases Brief Rep [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel BL, Lee H, Deignan JL, Strom SP, Kantarci S, Wang X, Quintero-Rivera F, Vilain E, Grody WW, Perlman S, Geschwind DH Nelson SF, 2014. Exome sequencing in the clinical diagnosis of sporadic or familial cerebellar ataxia. JAMA Neurol 71: 1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn P, 2003. NTE: one target protein for different toxic syndromes with distinct mechanisms? Bioessays 25: 742–745. [DOI] [PubMed] [Google Scholar]
- Hein ND, Rainier SR, Richardson RJ Fink JK, 2010a. Motor neuron disease due to neuropathy target esterase mutation: enzyme analysis of fibroblasts from human subjects yields insights into pathogenesis. Toxicol Lett 199: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein ND, Stuckey JA, Rainier SR, Fink JK Richardson RJ, 2010b. Constructs of human neuropathy target esterase catalytic domain containing mutations related to motor neuron disease have altered enzymatic properties. Toxicol Lett 196: 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufnagel RB, Arno G, Hein ND, Hersheson J, Prasad M, Anderson Y, Krueger LA, Gregory LC, Stoetzel C, Jaworek TJ, Hull S, Li A, Plagnol V, Willen CM, Morgan TM, Prows CA, Hegde RS, Riazuddin S, Grabowski GA, Richardson RJ, Dieterich K, Huang T, Revesz T, Martinez-Barbera JP, Sisk RA., Jefferie C., Houlde H., Dattan MT., Fin JK., Dollfu H., Moor AT.Ahmed ZM, 2015. Neuropathy target esterase impairments cause Oliver-McFarlane and Laurence-Moon syndromes. J Med Genet 52: 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liang X, Li Y, Wang J, Zaneveld JE, Wang H, Xu S, Wang K, Wang B, Chen R Sui R, 2015. Comprehensive molecular diagnosis of 67 Chinese Usher syndrome probands: high rate of ethnicity specific mutations in Chinese USH patients. Orphanet J Rare Dis 10: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmoch S, Majewski J, Ramamurthy V, Cao S, Fahiminiya S, Ren H, MacDonald IM, Lopez I, Sun V, Keser V, Khan A, Stránecký V, Hartmannová H, Přistoupilová A, Hodaňová K, Piherová L, Kuchař L, Baxová A, Chen R, Barsottini OG, Pyle A, Griffin H, Splitt M, Sallum J, Tolmie JL, Sampson JR, Chinnery P, Banin E, Sharon D, Dutta S, Grebler R, Helfrich-Foerster C, Pedroso JL, Kretzschmar D, Cayouette M Koenekoop RK, 2015. Mutations in PNPLA6 are linked to photoreceptor degeneration and various forms of childhood blindness. Nat Commun 6: 5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K, Kobayashi F, Miwa M, Shindo K, Isozaki E, Ishiura H, Tsuji S Takiyama Y, 2015. Novel mutations in the PNPLA6 gene in Boucher-Neuhäuser syndrome. J Hum Genet 60: 217–220. [DOI] [PubMed] [Google Scholar]
- Langdahl JH, Frederiksen AL, Nguyen N, Brusgaard K Juhl CB, 2017. Boucher Neuhäuser Syndrome - A rare cause of inherited hypogonadotropic hypogonadism. A case of two adult siblings with two novel mutations in PNPLA6. Eur J Med Genet 60: 105–109. [DOI] [PubMed] [Google Scholar]
- Mühlig-Versen M, da Cruz AB, Tschäpe JA, Moser M, Büttner R, Athenstaedt K, Glynn P Kretzschmar D, 2005. Loss of Swiss cheese/neuropathy target esterase activity causes disruption of phosphatidylcholine homeostasis and neuronal and glial death in adult Drosophila. J Neurosci 25: 2865–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil E, Serrano L, Scoles D, Cunningham KE, Han G, Chiang J, Bennett J Aleman TS, 2019. Detailed retinal phenotype of Boucher-Neuhäuser syndrome associated with mutations in PNPLA6 mimicking choroideremia. Ophthalmic Genet 40: 267–275. [DOI] [PubMed] [Google Scholar]
- Patsi O, De Beaufort C, Kerschen P, Cardillo S, Soehn A, Rautenberg M Diederich NJ, 2018. A new PNPLA6 mutation presenting as Oliver McFarlane syndrome. J Neurol Sci 392: 1–2. [DOI] [PubMed] [Google Scholar]
- Rainier S, Albers JW, Dyck PJ, Eldevik OP, Wilcock S, Richardson RJ Fink JK, 2011. Motor neuron disease due to neuropathy target esterase gene mutation: clinical features of the index families. Muscle Nerve 43: 19–25. [DOI] [PubMed] [Google Scholar]
- Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, Delaney C, Richardson RJ, Albers JW, Matsunami N, Stevens J, Coon H, Leppert M Fink JK, 2008. Neuropathy target esterase gene mutations cause motor neuron disease. Am J Hum Genet 82: 780–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Hein ND, Wijeyesakere SJ, Fink JK Makhaeva GF, 2013. Neuropathy target esterase (NTE): overview and future. Chem Biol Interact 203: 238–244. [DOI] [PubMed] [Google Scholar]
- Salgado P., Carvalho R., Brandão AF., Jorge P., Ramos C., Dias D., Alonso I.Magalhães M, 2019. Gordon Holmes syndrome due to compound heterozygosity of two new PNPLA6 variants - A diagnostic challenge. eNeurologicalSci 14: 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador F, Garcia-Arumi J, Corcostegui B, Minoves T Tarrus F, 1995. Ophthalmologic findings in a patient with cerebellar ataxia, hypogonadotropic hypogonadism, and chorioretinal dystrophy. Am J Ophthalmol 120: 241–244. [DOI] [PubMed] [Google Scholar]
- Sogorb MA, Pamies D, Estevan C, Estévez J Vilanova E, 2016. Roles of NTE protein and encoding gene in development and neurodevelopmental toxicity. Chem Biol Interact. 259: 352–357. [DOI] [PubMed] [Google Scholar]
- Song Y, Wang M, Mao F, Shao M, Zhao B, Song Z, Shao C Gong Y, 2013. Knockdown of Pnpla6 protein results in motor neuron defects in zebrafish. Dis Model Mech 6: 404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM, Andorf JL, Whitmore SS, DeLuca AP, Giacalone JC, Streb LM, Braun TA, Mullins RF, Scheetz TE, Sheffield VC Tucker BA, 2017. Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease. Ophthalmology 124: 1314–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M, Gonzalez MA, Lourenco CM, Coutelier M, Haack TB, Rebelo A, Hannequin D, Strom TM, Prokisch H, Kernstock C, Durr A, Schöls L, Lima-Martínez MM, Farooq A, Schüle R, Stevanin G, Marques W Jr. Züchner S, 2014. PNPLA6 mutations cause Boucher-Neuhauser and Gordon Holmes syndromes as part of a broad neurodegenerative spectrum. Brain. 137: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Synofzik M, Hufnagel RB Zuchner S 2015. PNPLA6-Related Disorders. In GeneReviews((R))(Eds Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K and Amemiya A). Seattle (WA): University of Washington, Seattle [Google Scholar]
- University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved. [Google Scholar]
- Tarnutzer AA, Gerth-Kahlert C, Timmann D, Chang DI, Harmuth F, Bauer P, Straumann D Synofzik M, 2015. Boucher-Neuhäuser syndrome: cerebellar degeneration, chorioretinal dystrophy and hypogonadotropic hypogonadism: two novel cases and a review of 40 cases from the literature. J Neurol 262: 194–202. [DOI] [PubMed] [Google Scholar]
- Teive HAG, Camargo CHF, Sato MT, Shiokawa N, Boguszewski CL, Raskin S, Buck C, Seminara SB Munhoz RP, 2018. Different Cerebellar Ataxia Phenotypes Associated with Mutations of the PNPLA6 Gene in Brazilian Patients with Recessive Ataxias. Cerebellum 17: 380–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topaloglu AK, Lomniczi A, Kretzschmar D, Dissen GA, Kotan LD, McArdle CA, Koc AF, Hamel BC, Guclu M, Papatya ED, Eren E, Mengen E, Gurbuz F, Cook M, Castellano JM, Kekil MB, Mungan NO, Yuksel B Ojeda SR, 2014. Loss-of-function mutations in PNPLA6 encoding neuropathy target esterase underlie pubertal failure and neurological deficits in Gordon Holmes syndrome. J Clin Endocrinol Metab 99: E2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiethoff S, Bettencourt C, Paudel R, Madon P, Liu YT, Hersheson J, Wadia N, Desai J Houlden H, 2017. Pure Cerebellar Ataxia with Homozygous Mutations in the PNPLA6 Gene. Cerebellum 16: 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winrow CJ, Hemming ML, Allen DM, Quistad GB, Casida JE Barlow C, 2003. Loss of neuropathy target esterase in mice links organophosphate exposure to hyperactivity. Nat Genet 33: 477–485. [DOI] [PubMed] [Google Scholar]
- Yoon G, Baskin B, Tarnopolsky M, Boycott KM, Geraghty MT, Sell E, Goobie S, Meschino W, Banwell B Ray PN, 2013. Autosomal recessive hereditary spastic paraplegia-clinical and genetic characteristics of a well-defined cohort. Neurogenetics 14: 181–188. [DOI] [PubMed] [Google Scholar]
- Yu SI, Kim JL, Lee SG, Kim HW Kim SJ, 2008. Ophthalmologic findings of Boucher-Neuhauser syndrome. Korean J Ophthalmol 22: 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccheo O, Dinsdale D, Meacock PA Glynn P, 2004. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 279: 24024–24033. [DOI] [PubMed] [Google Scholar]
- Zheng R, Zhao Y, Wu J, Wang Y, Liu JL, Zhou ZL, Zhou XT, Chen DN, Liao WH Li JD, 2018. A novel PNPLA6 compound heterozygous mutation identified in a Chinese patient with BoucherNeuhauser syndrome. Mol Med Rep 18: 261–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Yao F, Han X, Li H, Yang L Sui R, 2017. Rep1 copy number variation is an important genetic cause of choroideremia in Chinese patients. Exp Eye Res. 164: 64–73. [DOI] [PubMed] [Google Scholar]