Figure 2. Structure of Drosophila melanogaster Brat NHL domain bound to RNA.
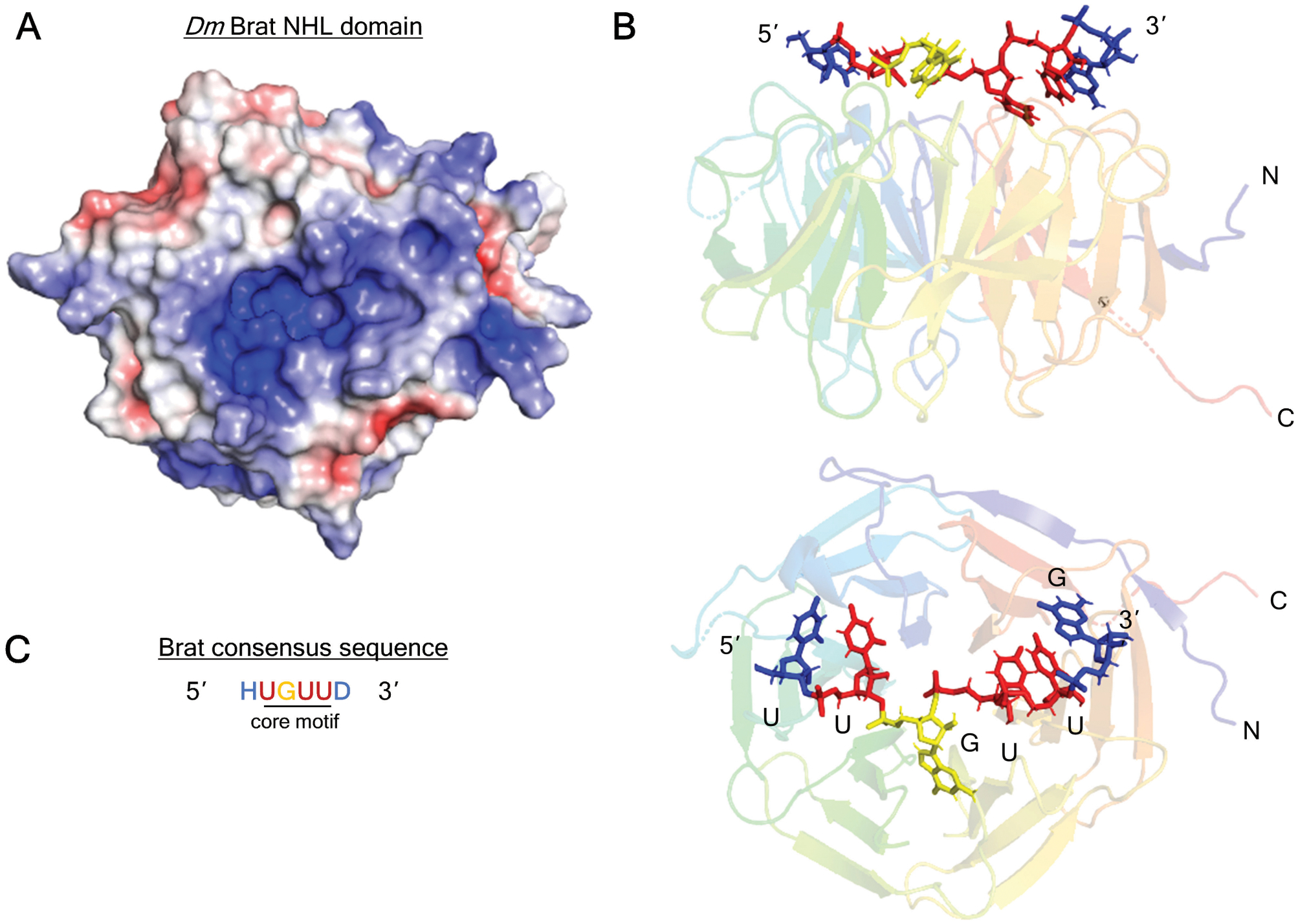
A. Positively-charged channel on the top, RNA-binding surface of Brat NHL domain (PDB: 1Q7F) (Edwards et al., 2003). The surface charge is visualized with Adaptive Poisson-Boltzmann Solver (APBS) macromolecular electrostatics calculation plugin in PyMOL. Blue denotes positive charge, red denotes negative charge.
B. Structure of the Brat NHL domain in complex with its RNA binding site from the hunchback mRNA 3’UTR (PDB: 4ZLR) (Loedige et al., 2015). The linear chain of nucleotides contact side chains of loop residues on the NHL surface.
C. Brat (and NCL-1) consensus binding motif consists of a core 5’-UGUU, with variable flanking nucleotides: H = (A/C/U), D = (A/G/U). The Brat binding motif consensus is derived from Loedige (2015) and Laver (2015).
