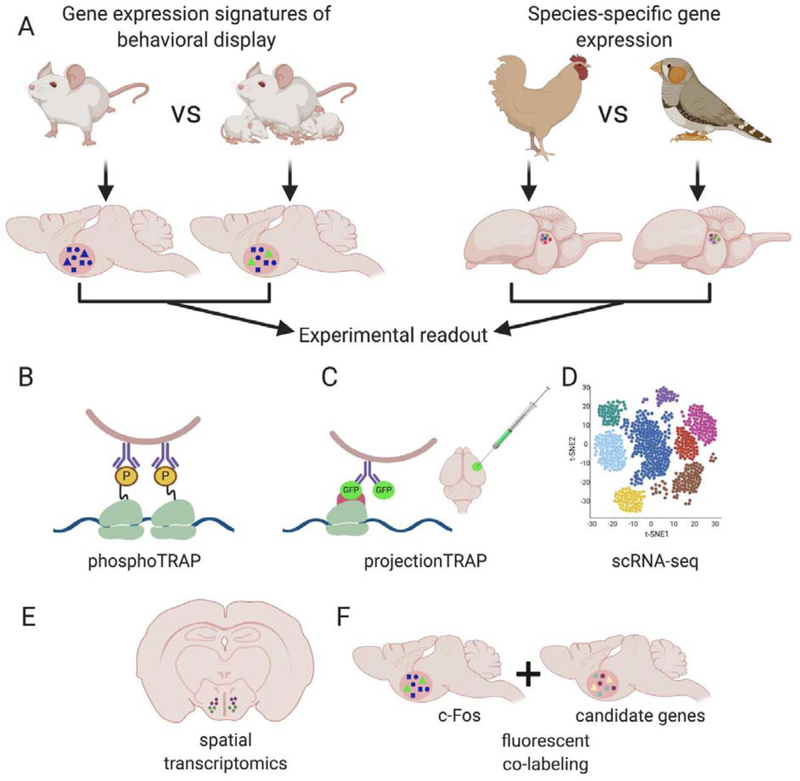
Figure 1. Strategies for pinpointing behaviorally-relevant genes.
(A) Schematic of behavioral paradigms that can be used to discern the genetic and molecular signatures of social behaviors. On the left, a mouse is shown with pups and no pups. Since it is known that the medial preoptic area (mPOA) is involved in parental behavior in mice, eliciting this behavioral response can be used to find cells in the mPOA that participate in the neural circuit involved in parental behavior display. This in turn would allow assessment of gene expression changes in specific cells that are relevant to parenting behavior. On the right, two species of birds with innate differences in behavior are shown; a vocal-learner songbird and a non-vocal learner bird (chicken). In this case, it is known that the medial portion of the dorsolateral thalamus (DLM) is one of the regions involved in vocal learning in birds (Goldberg and Fee 2011). Comparing both species would help identify key differences in the genetic profile of cells in the DLM of vocal learners and non-vocal learners. It could be the case, for example, that there are different quantities of a certain cell type in one species, similar cell types with different connectivity patterns, and/or specific genes with differential gene expression patterns.
(B-F) A summary of molecular approaches that can be employed to study social behaviors are shown.
(B) Phosphorylated Translating Ribosome Affinity Purification (phosphoTRAP) can be used to select for transcripts of genes being expressed during a behavioral response.
(C) ProjectionTRAP can be employed to capture transcripts of cells with specific projection patterns within a neural circuit, which is ideal for comparing projection pattern differences between species.
(D) Single cell RNA-seq (scRNA-seq) can be used in combination with TRAP methods to genetically profile relevant cells to a social behavior’s neural circuit.
(E,F) Information obtained from scRNA-seq can be further used for spatial transcriptomics
(E) and candidate gene co-labeling with c-Fos (F), to obtain an integral understanding of relevant genes being expressed in their native context. Figure created with BioRender.com.