Figure 3.
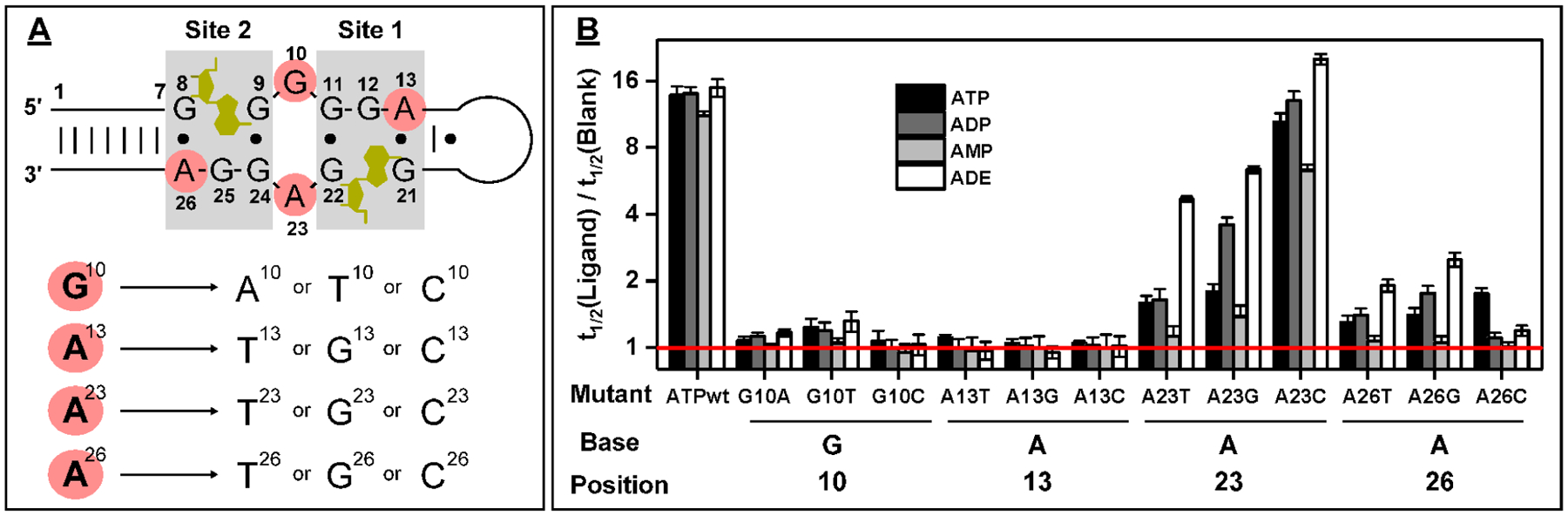
Design and characterization of ATPwt mutants. (A) Secondary structure of ligand-bound ATPwt, with mutated nucleotides highlighted in red. The ligand is highlighted in gold, and nucleotide positions are marked. Twelve different point mutants were generated by changing G10, A13, A23, A26 to either of the three alternative nucleobases. (B) The t1/2 ratio (with log2 scale) for each mutant in the presence of ATP, ADP, AMP, or ADE. The red line indicates a t1/2 ratio of 1, reflecting no binding-induced inhibition of enzyme digestion.
