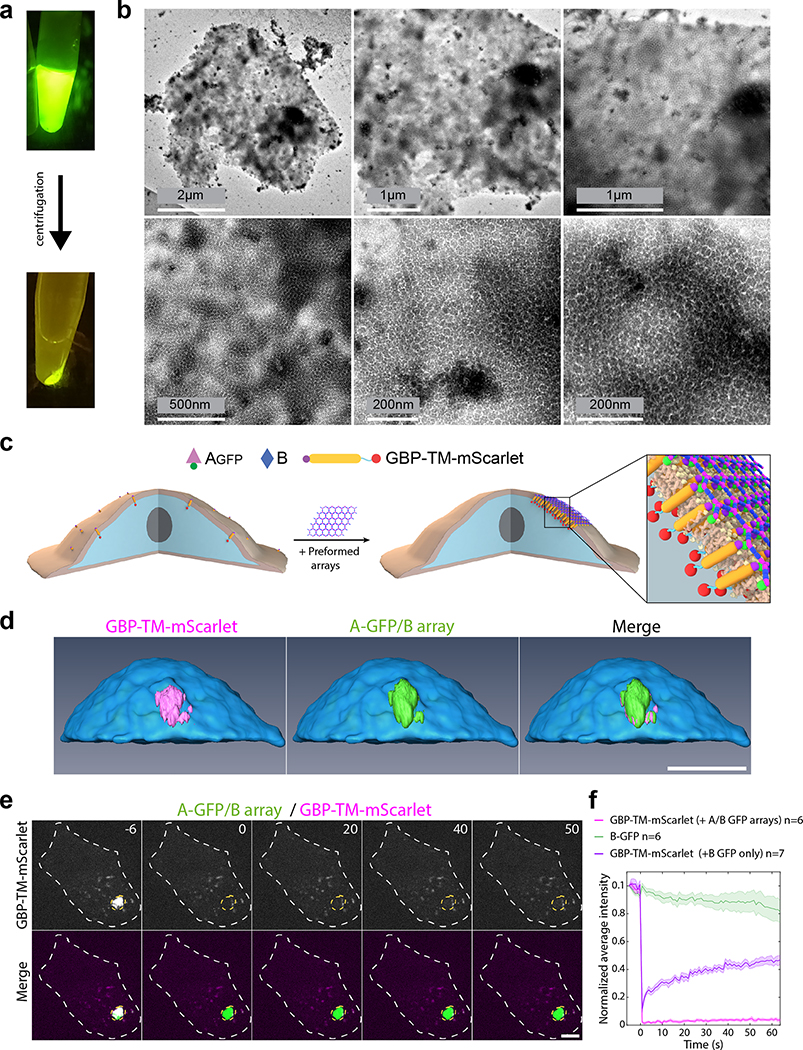
Extended Data Figure 6. Preformed arrays cluster transmembrane proteins in stable assemblies.
(a-b) Preformed arrays clusters characterization. (a) 2D arrays formed in-vitro by mixing Agfp+B in equimolar concentration (5uM) in buffer (25mM Tris-HCl, 150mM NaCl, 5% glycerol) supplemented with 500mM imidazole followed by overnight incubation at room temperature in eppendorf tube (total volume of 200μL). After polymerisation, solution is centrifuged, supernatant is discarded, and pellet is resuspend the same buffer. (b) Negative stain TEM images of the resuspended array pellet (10-fold dilution, see methods). (c-d) Clustering of transmembrane proteins by preformed arrays. (c) principle of the experiment: NIH/3T3 cells expressing GBP-TM-mScarlet are incubated with Agfp+B arrays for 30min leading to clustering of the mScarlet construct. This is the same scheme as in Fig. 3a reproduced here for clarity. (d) After incubation with preformed arrays, live cells are processed for imaging by spinning disk confocal microscopy. 3D z-stacks are acquired (11 μm, Δz=0.2 μm) and processed for 3D reconstruction. Note that the intracellular mScarlet protein signal overlaps perfectly with the extracellular GFP signal of the array. (e-f) mScarlet constructs clustered by the arrays are not dynamic. (e) Cells were incubated with Agfp+B arrays for 1 hour at 37°C, then the mScarlet signal was bleached and its fluorescence recovery monitored. The GFP signal was used to delineate the bleaching area. (f) Quantification of the effect seen in a (see methods). The mScarlet signal (magenta curve) does not recover, suggesting that GBP-TM-mScarlet molecules are stably trapped by the Agfp+B array. As a control that binding of Agfp alone (that is, not in an array) does not affect fluorescence recovery of GBP-TM-mScarlet (meaning that the array does not recover because all the GBP-TM-mScarlet is trapped by the Agfp+B array), we also performed FRAP experiments of GBP-TM-mScarlet in cells incubated with Agfp alone (purple curve). As expected, these recovers. Scale bars: (d) 12 μm; (e) 6 μm.