Extended Data Fig. 7. Altered H3K36me2 and H3K27me3 distribution in H1c/e-deficient GCB-cells.
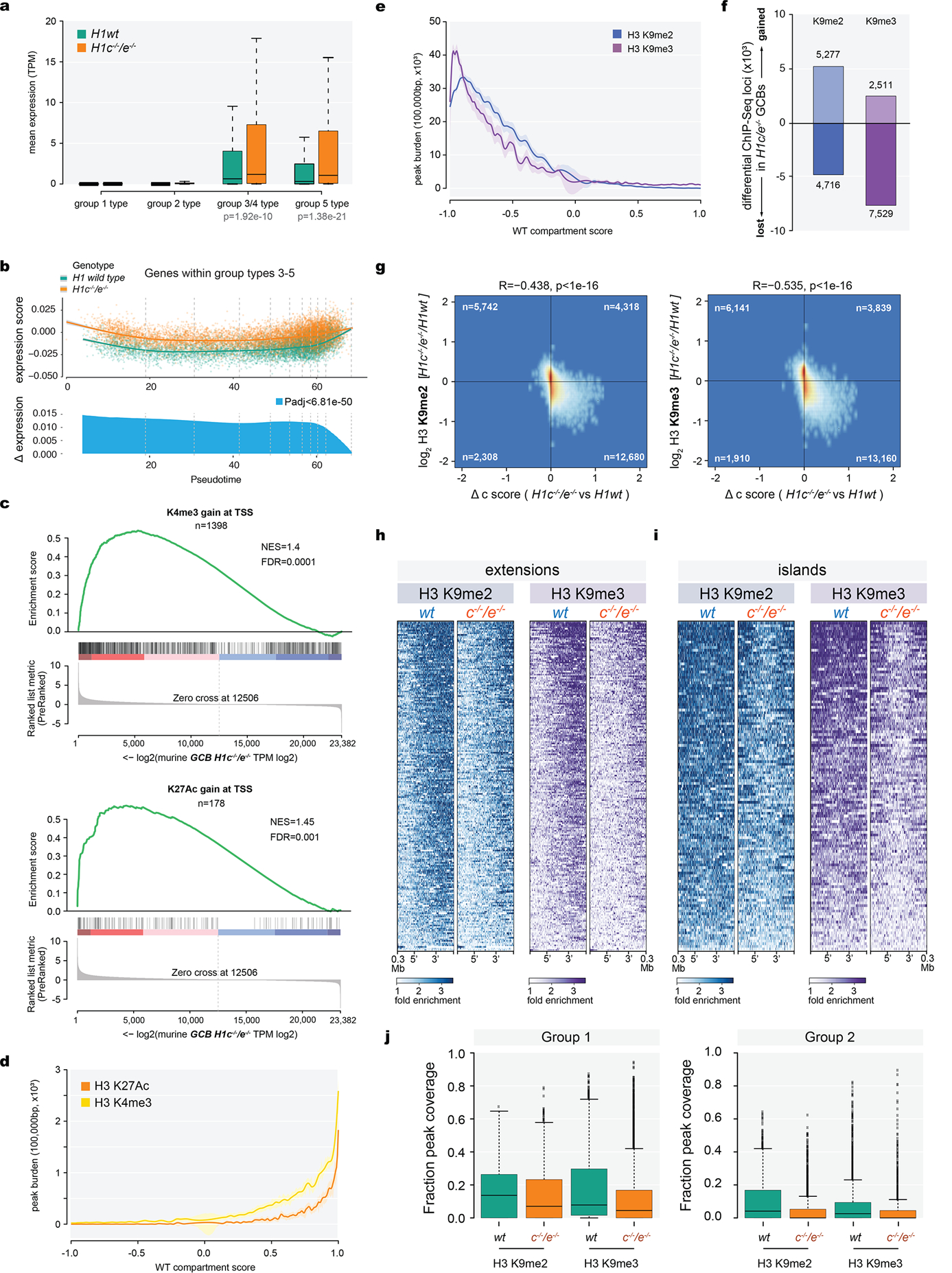
a-b, Mass spectrometry of H3 K36 (a) and K27 (b) post-translational modifications, log2-transformed and normalized to average wild type peak area from H1c−/−/e−/−(n=5) and WT (n=7) acid-extracted samples from GCB-cells; two-sided unpaired t-test: K36 unmod, ***P=0.0005; K36me1, ***P=0.0003; K36me2 ***P=0.0002; K36me3, P=0.93; K36ac, P=0.56; K27unmod, *P=0.0157; K27me1, **P=0.0072; K27me2, *P=0.0175; K27me3, ***P=0.0007; K27ac, P=0.9337. box plots show median and 25thto 75thpercentile, whiskers indicate data range; Data are representative from two independent experiments. c, Immunoblots for H1 (D4J5Q and AE-4 antibodies), H3K36me2, H3K27me3, EZH2, and NSD2 from sorted wild-type and H1c−/−/e−/−GC B-cells. Direct blue stain is included as loading control. A representative image of at least three experiments is shown. Uncropped gels are shown in Supplementary Figure 1. d, Mass spectometry-based relative abundance of H3.1/.2 (replication-dependent) and H3.3 (replication-independent) isoforms, shown as average % total peak area of H3 K27-K36 peptide containing H3.3-specific S31, in acid-extracted histones from WT (H3.3, 15.76%, n=7) and H1c−/−/e−/− (H3.3, 15.07%, n=5) GCB-cells; two-sided unpaired t-test, P=0.0004, Data are mean ± SD. e, Mass spectrometry of H3 K36 (top) and K27 (bottom) post-translational modifications across H3.1/2 (left) and H3.3 (right) isoforms, log2-transformed and normalized to average wild type peak area from samples acid-extracted from wild type (n=7) and H1c−/−/e−/−(n=5) GC B-cells; Two-sided unpaired t-test: H3.1/2 K36 unmod, ***P=0.0005; H3.1/2 K36me1, ***P=0.0003; H3.1/2 K36me2, ****P<0.0001; H3.1/2 K36me2, P=0.88; H3.1/2 K27unmod, *P=0.0100; H3.1/2 K27me1, *P=0.0162; H3.1/2 K27me2, *P=0.0129; H3.1/2 K27me3, ***P=0.0002; H3.3 K36unmod, ****P<0.0001; H3.3 K36me1, *P=0.036; H3.3 K36me2, P=0.15; H3.3 K36me3, P=0.5974; H3.3 K27unmod, P=0.1187; H3.3 K27me1, P=0.4743; H3.3 K27me2, P=0.1199; H3.3 K27me3, P=0.0628. Box plots show median and 25thto 75thpercentile, whiskers indicate data range. f, Unsupervised hierarchical clustering analysis of ChIP-seq data for H3K27me3 and H36me2 in biological triplicates from sorted H1c−/−/e−/−and wild-type H1GC B-cells.g, Genome wide correlation plot of log2 fold (H1c−/−/e−/−vs wild-type H1) change of normalized reads within ChIP-Seq peak union for H3K36me2 and H3K27me3. (Pearson correlation coefficient R=−0.453, P<1e-16). h, Heatmap of HiC compartment score, H3K36me2 and H3K27me3 centered within shifting B to A compartments (100kb) and surrounding 300kb for H1c−/−/e−/−and wild-type GC B-cells for compartment “extensions” (top). i, Fraction of ChIP-Seq peak coverage (H3K36me2 in red and H3K27me3 in blue) within 100kb compartments across HiC compartment score (X axis, −1 to 1) for WT GCB-cells. Cubic smoothing spline of data is presented with error bars as shaded regions indicate 99% confidence intervals. j, Fraction peak (H3K27me3 and H3K36me2) coverage of regions within shifting compartment groups 1–5 in H1c−/−/e−/−and wild-type H1 GC B-cells. Paired Wilcoxon test, Group 1: H3K27me3, P<1e-16; H3K36me2, P=0.597;Group 2: H3K27me3, P<1e-16; H3K36me2, P<1e-16; Group 3: H3K27me3, P=3.38e-13; H3K36me2, P<1e-16;Group 4: H3K27me3, P=8.71e-15; H3K36me2, P<1e-16;Group 5: H3K27me3, P<1e-16; H3K36me2, P<1e-16. Boxplot center represents median, bounds of box are 1stand 3rdquartile and whiskers extend out 1.5*interquartile range from the box. k,Scatter plot of H3K27me3 peak log2 fold change(H1c/e deficient compared to WT GCB-cells) versus WT compartment score for decompacting group 2. Gain of H3K27me3 (red dots) largely occurred within regions shifting from compartment B while loss of H3K72me3 (blue dots) were more prevalent within regions shifting compartments from compartment A.
